What a tangled web we may have weaved
A few words on spider butts, (self-assembling) hydrogels and proteinaceous obstructions discovered in autopsies
A paper was published back in August 2022 entitled: “Spidroin N-terminal domain forms amyloid-like fibril based hydrogels and provides a protein immobilization platform”.1 It was published by a group out of Sweden and involves an investigation into the usefulness of spider silk proteins as scaffolding for biotech purposes.
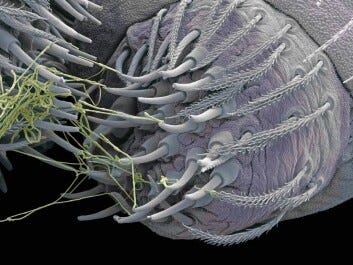
The reason I am bringing this paper up now (and summarizing it) is because it was recently linked to our preprint about DNA contamination in the COVID modified mRNA products by a follower on Twitter. The reason it was linked was because of the mystery open reading frame (ORF) discovered in the plasmids snapgened by Kevin McKernan, and also discussed in the publication entitled: “Are There Hidden Genes in DNA/RNA Vaccines?”.2
ORFs are spans of DNA between start and stop codons. Start and stop codons are the triplets of bases (in messenger RNA) that command the commencement and ending of translation in a protein. Whatever that protein may be.
So what’s the problem, you ask? Well, it’s not just that this ORF is absolutely not meant to be in the plasmid, just like the DNA is not meant to be in the vials. It’s not just that this ORF was absolutely not disclosed by Pfizer in their plasmid map, just like the SV40 elements were not disclosed.
WHO and FDA guidelines exist to prevent biological whoopsies like inclusion of superfluous potentially ‘active’ ORFs that may end up in in vivo contexts. The following paragraph is an excerpt taken verbatim from the WHO Technical Report Series No 941, 2007 Guidelines for assuring the quality and nonclinical safety evaluation of DNA vaccines.
…a DNA sequence homology check of the plasmid with the international databases (e.g. the National Center for Biotechnology Information, National Institute for Health, USA, and/or other international nucleotide databases) should be performed to investigate the presence of unintended sequences of biological significance such as those encoding cellular growth functions or alternative and unanticipated reading frames.3
These guidelines exist for very good reasons that go far beyond optimizing plasmids for use in vaccine component production: they exist because we really want to prevent mystery ORFs in biological product settings because of the risk of translatable ‘undesirable’ proteins (ahem - like amyloids) that may have unintended clinical outcomes in the in vivo setting (aka: adverse events). This particular ORF that Kevin found is exceedingly odd in that it is the antisense of the spike itself, although it has no known function as a protein.
The punchline is that when Kevin BLASTed this ORF in UniProt, he found that it has weak sequence homology (25.3%) to Major ampullate spidroin 1 MaSp1. The source organism listed is a bacterium called Nitrospirillum amazonense Y2. This MaSp1 is one of 2 types (the other one is MaSp2) used to make a specialized type of spider silk which is also the crux element of the paper.
Two open questions on our minds: Are superfluous whoopsie proteins being made in some people who were injected with the modified mRNA COVID-19 products? Do the vials contain DNA contaminant-associated amyloid-encoding genetic material?
I’ve already written extensively on the amyloidogenic nature of the spike protein but maybe this is yet another way for the circulatory system to get ‘gummed up’?
On spidroins
Spidroins are not an 80s band from Toronto but, in fact, spidroins are proteins used to make spider silk.
Interesting tidbit: You can’t farm spider silk like you can silkworm silk because spiders tend to eat each other. Not very profitable as a business. Better to use Crispr-Cas to gene edit silkworms to make them make spider silk?4
Spidroins are part of a large group of proteins called scleroproteins that also includes other structural proteins like keratin and collagen.5 The major ampullate spidroins (MaSp1 and 2 - mainly consisting of highly repetitive glycine and alanine blocks6) so-called because they are secreted by the major ampullate gland in spiders, are thus used for creating Dragline silk for those spider thrill seekers who rapel across Europe, and spin webs, and fly, and… transferance of sex hormones?

Silk produced by females provides a transmission channel for male vibratory courtship signals, while webs and draglines provide a substrate for female sex pheromones.7
Amazingly, the role of male-produced silk in mating is ‘under-studied’.8
Spidroin is really important composition-wise in dragline silk because of its associated toughness and strength. However, there many different types of spider silk composed of different types of proteins, including spidroins, all with specific functions. Shown in Figure 3 on the left are the various types of silks used for web construction with a little black spider hanging from a Dragline. Each of these silks has their own protein compositions, including spidroin proteins, and originate from different glands of which some spiders, like Araneus diadematus, have 7. On the right is a proposed model for Dragline composition and structure involving various protein structures (including secondary structures: beta sheets and alpha helices) interconnected by an amorphous matrix.
![Schematic of the spider's orb web, structural modules, and spider silk structure.[15] On the left is shown a schematic drawing of an orb web. The red lines represent the dragline, radial line, and frame lines, the blue lines represent the spiral line, and the centre of the orb web is called the "hub". Sticky balls drawn in blue are made at equal intervals on the spiral line with viscous material secreted from the aggregate gland. Attachment cement secreted from the piriform gland is used to connect and fix different lines. Microscopically, the spider silk secondary structure is formed of spidroin and is said to have the structure shown on the right side. In the dragline and radial line, a crystalline β-sheet and an amorphous helical structure are interwoven. The large amount of β-spiral structure gives elastic properties to the capture part of the orb web. In the structural modules diagram, a microscopic structure of dragline and radial lines is shown, composed mainly of two proteins of MaSp1 and MaSp2, as shown in the upper central part. In the spiral line, there is no crystalline β-sheet region. Schematic of the spider's orb web, structural modules, and spider silk structure.[15] On the left is shown a schematic drawing of an orb web. The red lines represent the dragline, radial line, and frame lines, the blue lines represent the spiral line, and the centre of the orb web is called the "hub". Sticky balls drawn in blue are made at equal intervals on the spiral line with viscous material secreted from the aggregate gland. Attachment cement secreted from the piriform gland is used to connect and fix different lines. Microscopically, the spider silk secondary structure is formed of spidroin and is said to have the structure shown on the right side. In the dragline and radial line, a crystalline β-sheet and an amorphous helical structure are interwoven. The large amount of β-spiral structure gives elastic properties to the capture part of the orb web. In the structural modules diagram, a microscopic structure of dragline and radial lines is shown, composed mainly of two proteins of MaSp1 and MaSp2, as shown in the upper central part. In the spiral line, there is no crystalline β-sheet region.](https://substackcdn.com/image/fetch/$s_!TaEL!,w_1456,c_limit,f_auto,q_auto:good,fl_progressive:steep/https%3A%2F%2Fsubstack-post-media.s3.amazonaws.com%2Fpublic%2Fimages%2Fca7a3c0e-6632-4f39-8173-f109c9185a7c_900x381.png)
Inside the spider’s hindquarters (aka: spider butt), is the major ampullate gland as shown below in Figure 4. The Dragline silk is produced from spidroins that are stored in the major ampullate gland. It is a pipeline of decreasing diameter with chamber-associated specific physiological, physical and chemical characteristics, that allows for almost instant production of super strong/tough silk fibers by pulling, from the stored spidroin proteins.

The pulling/spinning is done by the spinnerets on the ass-end of the spider.

Dragline silk has incredibly high tensile strength and ductility (flexible) making it perfect as the spider’s bungy-jumping lifeline. This is why humans are so interested in copying it for repurposing as biomaterials. In fact, it is actually tougher than both kevlar and steel. It’s absolutely mind-blowing to me that Draglines also have torsional memory. This is why the spider doesn’t spin or twist when ‘hanging by a thread’.9
Not only are medical and biotech arenas trying to get in on the spider silk action, but the military are as well. For example, spider silk technology is used in the production of bullet-proof vests. You might also find it interesting that as far back as 2011, there have been published works on introducing recombinant silk proteins into mammals for ‘regenerative medicine’.1011
Furthermore, fibers developed from spidroin are tolerated in vitro, in cell culture, and in vivo, in animals like pigs, as no signs of either inflammatory response nor body reaction were shown to these fibers. These results suggest that they could be used in medicine without risk of biocompatibility issues and thus potentially lead to many new opportunities in tissue engineering and regenerative medicine.12
On N-terminal (NT) domains
N-terminal domains are one of the two bookends of each amino acid (remember there are 21 amino acids in eukaryotes) and effectively also bookend proteins or protein chains (polypeptides). They are called N-terminal because they involve the presence of a Nitrogen (N) atom in the form of an NH2. The other bookend is called the Carboxyl-terminus or C-terminal because they involve the presence of a Carbon (C) atom in the form of a COOH. Let’s refer to the N-terminus as the head and the C-terminus as the tail. Each amino acid is bonded (a covalent chemical bond called a peptide bond) from head to tail and thus, all protein chains and all proteins in general, will have an N and a C-terminal. If you think of an amino acid as a pearl, then a ‘string’ of pearls would be akin to primary protein structure whereby the string linking the pearls would be the peptide bonds formed between the amino acids via the N and C-terminals. When these strings are bonded by hydrogen bonds side-by-each, they are called beta sheets, and when they are coiled they are called alpha helices. These are the secondary protein structures that we all know and love.

For more introductory and specific context from the paper, the NT domains in the paper are alpha helical as depicted in Figure 7 (pdb - 4fbs).

On hydrogels
Hydrogels are 3-dimensional cross-linked fibrous structures held together either by chemical (covalent) or physical (hydrogen) bonds, electrostatic interactions, van der Waals forces, or physical entanglements.13
…Or if you like the Wikipedia definition:
A hydrogel is a biphasic material, a mixture of porous, permeable solids and at least 10% by weight or volume of interstitial fluid composed completely or mainly by water. In hydrogels the porous permeable solid is a water insoluble three dimensional network of natural or synthetic polymers and a fluid, having absorbed a large amount of water or biological fluids.14
Figure 8 shows a schematic demonstrating the formation of the cross-linking of self-assembled nanofibers. Basically, hydrogels are 3-D meshes of hydrophillic nanofibers bound together.

Hydrogels are used to make such glorious things as contact lenses15, biosensors and glues. They are inspired by natural hydrogels such as gelatin (derived from collagen). There is nothing so new about hydrogels in biotech and industry, and they have been being developed and used for decades, but applications and variations continue to expand.
Hydrogels are classified based on many factors such as the composition of the nanofibers (polymers) used, the way they are cross-linked, how they respond to stimuli, as shown in Figure 9, and their ionic charge.
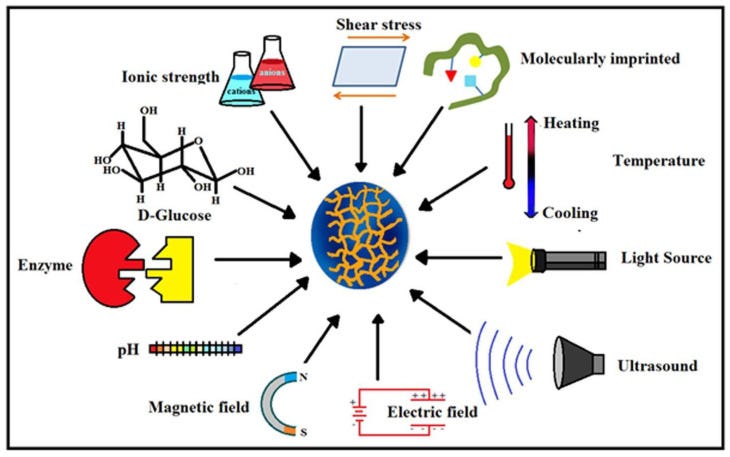
I highly recommend reading the papers entitled: “Fundamental Concepts of Hydrogels: Synthesis, Properties, and Their Applications”16 and “Hydrogels: Properties and Applications in Biomedicine”17 for comprehensive information pertaining to hydrogels.
In the abstract of the spidroin paper, the authors write:
Here, we report that recombinant miniature spidroins, and importantly also the N-terminal domain (NT) on its own, rapidly form self-supporting and transparent hydrogels at 37 °C.
…we find that NT and miniature spidroins undergo a structural transition into β-sheets and amyloid-like fibrils upon gel formation.
Figure 10 (Figure 8 from the paper) shows an overview of NT gelation. Gelation is the act of going into gel form. Interestingly, the NT samples incubated at pH 8 and 7 both gelled after 30 min at 37 °C but they differed by clarity: the pH 8 gel remained clear and the pH 7 gel was more opaque (shown in Figure 11). Figure 10 shows the NT alone and NT tagged with green fluorescent protein (GFP) and purine nucleoside phosphorylase (PNP). The main reason the authors got excited was that they were able to fuse other proteins to the recombinant only-NT domain and form hydrogels with the embedded proteins remaining functional. And again, these hydrogels comprise amyloid-like fibrils.


Some of the mysteries being unraveled in spider silk science include how spiders maintain spidroin solubity in the ampulla and also how they produce/spin the silk so fast ‘out of storage’. It turns out it has a lot to do with the changing physiological conditions in the glands of the spider that alter the stability and structures of the N and C-terminal domains of the spidroins.18
Part of the biotech of spidroin science involves looking at the effects of changing various physiological parameters like pH, and determining how this affects fiber production and quality. What the authors found from their studies was that when they incubated recombinant mini-spidroins at 37°C, they were able to form hydrogels whereby a structural transition from α-helix into β-sheets resulted in the formation of amyloid-like fibrils.
The finding that NT on its own could form gels was unexpected considering the common notion that NT is a highly soluble and stable protein and that previous reports of recombinant spidroin hydrogels have attributed the gelation effect to conformational changes in the repetitive region and/or CT.
I would like refer to a paper entitled: “Decoding the secrets of spider silk” by Eisoldt et al.19 to show a cartoon version of the journey of the spidroin proteins to assembled fibers from tail to exterior via the major ampulate gland. I think it is important to understand what the spidroins go through to end up as stable NT-dimerized β-sheet-rich proteins that enable fiber assembly. This is more for me to mentally compare the spider stuff with what they did in the paper.
You can see from this neat schematic that the spidroins start off as secreted proteins and get stored at very high concentrations (50%) in the ampulla with liquid crystalline properties. Spidroins form micellar-like structures controlled by their amphiphilicity to prevent irreversible aggregation. The micellar-structures undergo chemically and physically-induced changes in the duct and move through sequentially smaller diameter spaces with sequentially lower pHs to ultimately result in assembled fibers by removal of water, dimerization of the NT domains, beta sheet formation and protein alignment.20 This is shown in the bottom right corner.
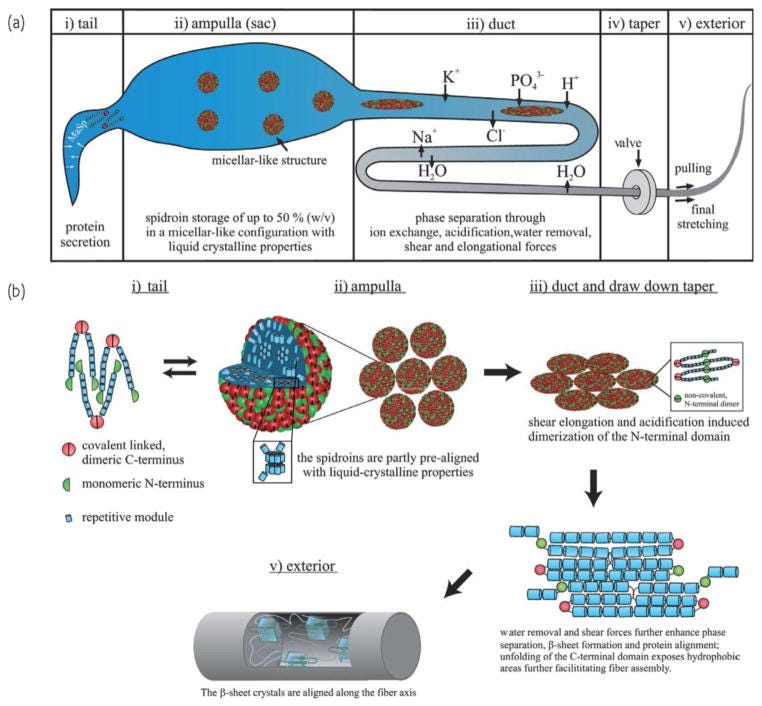
On the subject matter of the spider stuff, the authors of the spidroin paper write:
NT is monomeric under conditions that reflect those in the gland lumen and mediates solubility to the spidroins, but at decreased pH, protonation of a series of carboxylate side chains leads to NT dimerization with a pKa of around 6.5, which stabilizes NT and locks the spidroins in large networks. Thus, NT plays a key role in the silk formation process by transitioning from a monomer in the dope to a dimer in the fiber.
The important difference between the spider stuff21 and the recombinant lab stuff is the way in which the fibers form and it is kind of surprising, ie: α-helix dimerization and β-sheet formation/protein alignment in the former, and α-helix conversion into β-sheets in the latter using temperature as a means to induce gel formation.
Even though I feel I don’t quite yet understand the spider or the recombinant lab stuff, it is an amazing thing that people are doing with regard to the latter: making recombinant spidroins that form hydrogels. It is even more amazing that these synthetics can self-assemble into a variety of shapes, films or hydrogels.
I do highly recommend reading the decoding paper. It’s so awesome. They write:
So far, no biomimetic spinning process exists that would allow the formation of native-like spider silk fibers from recombinant silk proteins. However, despite the nonexistent spinning process most recombinant spider silks can self-assemble into non-natural shapes such as spheres, capsules, films, non-wovens or hydrogels (Fig. 5), which have a high application potential.

So what have we learned?
The spidroin paper is all about rapid formation of self-supporting and transparent hydrogels from recombinant spidroins - and NT on its own - in meh conditions. ‘The gelation is caused by NT α-helix to β-sheet conversion and formation of amyloid-like fibrils.’
So where does that leave us with our original question?
Are superfluous whoopsie proteins being made in some people who were injected with the modified mRNA COVID-19 products?
It might be the case. We need to find out.
Do the vials contain DNA contaminant-associated amyloid-encoding genetic material?
They might. We need to find out if any proteins and what proteins, are being passed to people and if these proteins are being translated. And if they are being translated, what are they being translated into?
Could it be that amyloidogenic peptides akin to spidroin are being translated in people and subsequently creating immobilizing hydrogel scaffolds for other proteins?
Comments welcome.
Arndt, T., Jaudzems, K., Shilkova, O. et al. Spidroin N-terminal domain forms amyloid-like fibril based hydrogels and provides a protein immobilization platform. Nat Commun 13, 4695 (2022). https://doi.org/10.1038/s41467-022-32093-7
Beaudoin et al., Are There Hidden Genes in DNA/RNA Vaccines? HYPOTHESIS AND THEORY article. Front. Immunol., 08 February 2022 Sec. Vaccines and Molecular Therapeutics. Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.801915
https://cdn.who.int/media/docs/default-source/biologicals/vaccine-standardization/dna-vaccines/annex-1-dna-vaccines.pdf (Page 65)
https://www.eurekalert.org/news-releases/1001587
https://en.wikipedia.org/wiki/Spidroin
Simmons, A. H.; Michal, C. A. & Jelinski, L. W. (1996). "Molecular orientation and two-component nature of the crystalline fraction of spider dragline silk". Science. 271 (5245): 84–87
https://en.wikipedia.org/wiki/Spider_silk
Scott, Catherine E.; Anderson, Alissa G.; Andrade, Maydianne C. B. (August 2018). "A review of the mechanisms and functional roles of male silk use in spider courtship and mating". The Journal of Arachnology. 46 (2): 173–206. doi:10.1636/JoA-S-17-093.1
Emile, O., Le Floch, A. & Vollrath, F. Shape memory in spider draglines. Nature 440, 621 (2006). https://doi.org/10.1038/440621a
Lazaris, A., Arcidiacono, S., Huang, Y., Zhou, J.-F., Duguay, F., Chretien, N., et al. (2002). Spider Silk Fibers Spun from Soluble Recombinant Silk Produced in Mammalian Cells. Science 295, 472–476. doi:10.1126/science.1065780
Rising A, Widhe M, Johansson J, Hedhammar M. Spider silk proteins: recent advances in recombinant production, structure–function relationships and biomedical applications. Cell. Mol. Life Sci. (2011) 68:169–184
https://en.wikipedia.org/wiki/Spidroin
https://www.sciencedirect.com/topics/engineering/hydrogel
https://en.wikipedia.org/wiki/Hydrogel
https://www.lensesonline.co.nz/blog/the-history-of-contact-lenses-from-glass-to-silicone-hydrogel/
Bashir S, Hina M, Iqbal J, Rajpar AH, Mujtaba MA, Alghamdi NA, Wageh S, Ramesh K, Ramesh S. Fundamental Concepts of Hydrogels: Synthesis, Properties, and Their Applications. Polymers (Basel). 2020 Nov 16;12(11):2702. doi: 10.3390/polym12112702. PMID: 33207715; PMCID: PMC7697203
Ho TC, Chang CC, Chan HP, Chung TW, Shu CW, Chuang KP, Duh TH, Yang MH, Tyan YC. Hydrogels: Properties and Applications in Biomedicine. Molecules. 2022 May 2;27(9):2902. doi: 10.3390/molecules27092902. PMID: 35566251; PMCID: PMC9104731
Rising A, Johansson J. Toward spinning artificial spider silk. Nat Chem Biol. 2015 May;11(5):309-15. doi: 10.1038/nchembio.1789. Epub 2015 Apr 17. PMID: 25885958
Lukas Eisoldt, Andrew Smith, Thomas Scheibel, Decoding the secrets of spider silk, Materials Today, Volume 14, Issue 3, 2011, Pages 80-86, ISSN 1369-7021, https://doi.org/10.1016/S1369-7021(11)70057-8
Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: spider silk. Acta Biomater. 2014 Apr;10(4):1612-26. doi: 10.1016/j.actbio.2013.08.020. Epub 2013 Aug 17. PMID: 23962644; PMCID: PMC3926901
Landreh, M. et al. Mass spectrometry captures structural intermediates in protein fiber self-assembly. Chem. Commun. 53, 3319–3322 (2017)


Jessica,
This is an incredible piece of writing. The science that you have outlined here is incredibly concerning and incredibly important. You did an excellent job of making very complex concepts accessible to the public. There is no question in my mind that this is more than enough to justify a worldwide moratorium on the modRNA shots and to launch disciplinary investigations into why Pfizer duped health regulatory agencies and why these regulatory agencies failed to do their basic homework in terms of quality control on behalf of the public they are supposed to serve.
Whoopsies Were NOT Made ;-)