RSFIEDLLFNKV... are we looking at weaponized amyloidosis?
And what of prions? An exploratory piece.
First I need to tell you guys that the number of Creutzfeldt-Jacob Disease (CJD)/prion disease reports in VAERS since the deployment of the COVID-19 injections has far surpassed the background rate for the U.S. for the year.
Before I go any further, I need to provide some serious background. Get a cup of coffee and settle in. This is a long one. I think that there’s no point in trying to convey this information properly unless I believe that my readers really understand some of the basics. (This has been an incredible learning experience for me and I hope I have this right so far.)
On proteins
Most people think of a steak when they hear the word protein. I used to. But proteins are more than steak. They are large biomolecules - energetically stable, compact, folded strings of amino acids whose folded structure determines their function. They do all sorts of things for us like metabolic reaction catalysis, DNA replication, providing cellular structure, molecular transport and so much more! You can purify them too! That’s what I did a lot of during my last Post Doc. Proteins are absolutely essential to life and living organisms. Short strings of amino acids are generally not termed proteins but peptides.

On prions
Among the many, many proteins1 that we humans express, we have prion proteins. Prion proteins or cellular prion proteins (PrPc), are native to humans, are in our brains and expressed all the time. When these particular proteins mis-fold, then, Houston, we have a problem.
These mis-folded proteins are called prions (mis-folded prion protein (PrPSc - Sc is for Scrapie2 3)) and are associated with neuro-degenerative diseases like Alzheimer’s disease. It seems that they can transform or ‘teach’ other prion proteins to mis-fold (transmissible), as well. It’s not really teaching as much as proximal disallowance of proper folding. This ‘enables’ an autocatalytic reaction whereby the PrPSc catalyzes the mis-folding of PrPc to produce more PrPSc due to the fact that prions are resistant to proteolysis - the process of ‘removal’ of mis-folded proteins.
Only prion proteins can mis-fold to produce prions and, again, these prions are special in that they are resistant to proteolysis. It is important to note that all proteins go through the folding process (as shown on the left-hand-side of the left screenshot in Figure 3) and if and when mis-folds occur, either chaperones guide these mis-folded proteins to ultimate final proper folding or these mis-folded proteins get eliminated by proteases.4 Imagine how long we would last if these brilliant maintenance and ‘house-keeping’ mechanisms weren’t in place, eh? Mis-folded proteins abound! Even so, this ‘house-keeping’ doesn’t work for prion proteins. Which makes me wonder why the hell we have them in the first place. But, that’s for another day.
Back to VAERS
There are 59 reports of CJD and prion diseases in VAERS to date (June 10, 2022). This is frightening. The background rate of CJD in the United States is 1/1M per year. Since the population of the U.S. as of June 6, 2022 is ~334M5, then we would expect no more than 334 cases of CJD reported. If we consider an extraordinarily low under-reporting factor (URF)6 of 6, then we have already surpassed the background expected number of cases reported by 20 reports. If we use the calculated URF of 31 as referenced in (2), then we have 1,829 cases of prion diseases (CJD) in the U.S., to date. And this is just data from VAERS. And this is just in the context of the COVID-19 shots. ~589M7 doses of the COVID-19 injections (Moderna, Pfizer and Janssen) have been doled out in the States as of June 6, 2022. This means that we are seeing ~3.1 incidents of prion diseases/CJD per million Americans - 3 times the background rate. Already.

The paper that scares me
I spent all day reading the papers linked to this article written by Walnut (Walter Chestnut), and they are fascinating. One in particular caught my eye. In summary, the idea here is that a conserved coronavirus spike protein peptide forms amyloid nano-structures and hydrogels under pH-dependent (pH = 4) conditions and that this has applications in materials science for slow release drug delivery.
A previous Substack I wrote covered the subject of the SARS-nCoV-2 spike having amyloidogenic peptides derived from FKNIDGYFKIYSKHTPINLV (say that 5 times fast). You can read that Substack here. This is the same, but different, from that. Both papers investigate functional aspects of spike peptides pertaining to amyloid formation. I also wrote a Substack on prion diseases. You can read that here.
Please refer to the paper entitled: “Amyloid and Hydrogel Formation of a Peptide Sequence from a Coronavirus Spike Protein” published January 4, 2022 in ACS Nano by Hamley and Castelletto. Now, for all intents and purposes, this is a materials science paper, but to me, it raises some serious Nancy Pelosi eyebrows.
A series of experiments were done using a coronavirus spike-derived peptide RSAIEDLLFDK(V). They found that this coronavirus spike peptide has really interesting properties under specific pH conditions that aren’t predicted by amyloid aggregation algorithms. A sequence-similar peptide, RSFIEDLLFNKV, is found in the SARS-nCoV-2 spike protein. So what I want to know is: does an analogous spike protein peptide in the SARS-nCoV-2 spike and/or the modified mRNA that encodes the spike protein in the injections result in the formation of nanotapes and amyloids as well under certain chemical/physiological conditions?
Visualizing protein structure to try to understand paper implications
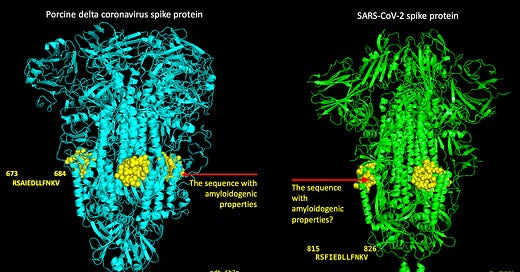
Visualizing protein structure using X-ray crystallography or cryo-electron microscopy, for example, is important. Visualization of a protein’s structure, and where particular sequences of amino acids ‘reside’, can give us insight on functional aspects of both the protein as a whole, and of particular sequences of amino acids in the protein. For example, sequences on the surfaces of proteins are probably located in accessible places, like the surface, because they contain a binding site for some cognate protein. The following diagram (Figure 7) shows the sequences of amino acids on the SARS-nCoV-2 spike protein that comprise the receptor binding domain (RBD). This is the SARS-CoV-2 Spike protein trimer with the RBD in the ‘UP’ position (pdb 6X2A) and the yellow box outlines the RBD that’s ‘UP’ and ready for binding ACE-2. I blacked out 2 out of 3 of the trimeric structures to make it easier to see.

With regard to the peptides RSAIEDLLFNKV and RSFIEDLLFNKV, I added some yellow spheres to the crystal structures for the porcine and the SARS-nCoV-2 spikes to visualize where these sequences of amino acids reside on the structures as shown in Figure 8. I did this by aligning the pork spike to the SARS-nCoV-2 spike in Pymol and then highlighting the amino acid sequences. Somewhere, Homer is drooling and saying, ‘MMMMM. Pork spike’.

Let’s now go to Figure 4 in their paper where they show transmission electron microscopy (TEM) images of the peptide RSAIEDLLFDK(V) (from a coronavirus spike protein - supplied by Peptide Synthetics) at different pHs (3-12). pH, just in case you forgot your basic chemistry (like I always do), is basically just a difference in hydrogen ion concentration represented on a log scale: higher hydrogen concentration = more acidic (acidic vs. alkaline). For some context, battery acid has PH = 0, stomach acid has pH = 1.5-2.0, tomato juice, acid rain and lysosomes have PH = 4-4.5, pure water has PH = 7 and liquid drain cleaner has PH = 14. Stay away from battery acid and liquid drain cleaner.

When they dissolved samples of the peptide (RSAIEDLLFDK) in acidic solution (PH 3) they didn’t see the fibrillar structures (top left). Instead, they saw what they referred to as ‘globular structures’ that are consistent with disordered (peptide) monomers in solution. When they increased the pH just by 1 to pH = 4, they saw amyloid structures or fibrils. They refer to these structures as extended amyloid structures or nanotapes. When they increased the pH to 5, they saw the peptide self-assemble ‘into wide nanotapes, up to 100 nm thick, which in some areas unbundle into ∼20 nm thick nanotapes’, as shown in the top right square. In a nutshell, they kept increasing the pH incrementally to see what effect it had on the formation of these fibrils or nanotape structures and saw clearly that there was a sweet spot for nanotape formation at pH = 4-5.
So let’s back-up. They claim in the very first sentence of the paper that:
The sequence RSAIEDLLFDKV is found in many coronaviruses including the human common cold coronavirus spike protein as well as other coronavirus spike proteins from other animals.
I thought a little about the implications of this. Again, this is a materials science paper and they really care about the potential applications for ‘pH-triggered gels and/or for slow release applications’. But, what I still don’t know is: does the modified mRNA in the Pfizer and Moderna injections encode amyloidogenic peptides that result in the formation of nanotapes and amyloids under certain chemical/physiological conditions? And this brings me back to one of the most important questions I posed well over a year ago and that is: ‘What are the resultant peptides of the modified mRNA following translation?’ (I Substacked about this (of course) and you can read that here.)
If the modified spike mRNA is not translated into its complete product by the host cells, and rather into incomplete ‘bits’ or peptides, then are any of these peptides amyloidogenic? Is it possible that one of these peptides is the analog peptide discussed here? Does that analog peptide (the conserved coronavirus SARS-nCoV-2 spike protein peptide) form amyloid nano-structures and hydrogels under pH-dependent (pH = 4) conditions? If so, where in the body could this happen? Could it happen? Stomach acid (pH = ~4)? Lysosomes (pH = ~4.5)8?
Just to be clear, it has been shown repeatedly in the literature that prion-like domains or peptides exist in the spike protein of SARS-nCoV-2 and these are specific to SARS-2.9 So again, what of our modified spike?
Some more scary facts
Proteins can convert from alpha helices to beta sheets thus exposing hydrophobic amino acids to promote protein aggregation. Just to remind everyone, alpha helices and beta sheets are both secondary structures with regard to protein organization.
Moreover, non-native beta sheet structures are common in amyloid fibrils formed by a variety of pathogenic and even non-pathogenic proteins and peptides. In all of these cases, the formation of alpha helix precedes the appearance of beta sheet, which suggests that conversion from the simpler, more local helix structure to the often more convoluted sheet architecture during folding and pathogenic mis-folding processes could be a unifying principle of general importance.
I am sorry to do this, but I looked at the Wikipedia site for prions (updated 2 weeks ago) and they have a paragraph about Weaponization. Of course they do. Where would humans be without paranoid psychopaths turning everything in to a weapon.
“Silly monkeys, give them thumbs, they make a club and beat their brother down.” “Silly monkeys, give them thumbs, they forge a blade, and where there’s one, they’re bound to divide it.” Maynard James Keenan
My closing thoughts
The prion thing is not the same thing as the amyloid thing, but they may be more inter-linked that we think. I don’t know yet. Walnut brings up the concept of a ‘prioloid’ in his Substack. I don’t know what I think of this yet. The one thing I know is that prions aggregate due to their high beta sheet compostion and are histochemically and ultrastructurally identical to amyloid.[2]
It’s time to talk solutions. And you should read these too.10 11 12 13
If some of the peptides in the SARS-nCoV-2 spike protein are able to form amyloid plaques then we will need treatment for those affected. Even more horrifying, if the modified mRNA (that was codon-optimized) contains amyloidogenic peptides that are able to form amyloid plaques, then the people injected are in serious trouble and will need some sort of solution. Well, a lot are dying now - the serious trouble has unfortunately passed for them. By the way, Sudden Adult Death Syndrome (SADS)? Really?
I looked up some fibrinolytic agents because I was asked by an MD the other day what to suggest to help a patient of his with recent diagnosis of amyloid plaques in the brain. I had no idea what to say. One person suggested in the comments section of Walnut’s Substack that Lumbrokinase nattokinase is something to look at. But it might involve eating dirt and earth worms.14
I will return my lovelies. If you want my advice, really learn how bad and irreversible amyloid plaque formation is (maybe just mention the word Alzheimer’s - Chris Masterjohn wrote a piece on Alzheimer’s a while ago and it is thorough and relevant) and convey to your loved ones being told they need to get injected again that the shots may cause them specific harms. Chris wrote that the accumulation of amyloid plaques are a universal hallmark of Alzheimer's but not necessarily the cause. Agreed.
The thing that worries me about this is that the systemic formation of amyloid plaques would explain so many, if not all of the severe adverse events that we are seeing in VAERS and other adverse event reporting systems. To me, it would explain myocarditis.
https://sandwalk.blogspot.com/2016/12/how-many-proteins-in-human-proteome.html
McKinley, M.P., Prusiner, S.B. (1987). Scrapie Prions, Amyloid Plaques, and a Possible Link with Alzheimer’s Disease. In: Altman, H.J. (eds) Alzheimer’s Disease. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-6414-0_7.
https://en.wikipedia.org/wiki/Scrapie
https://www.nature.com/scitable/topicpage/protein-structure-14122136/
https://www.worldometers.info/world-population/us-population/
Rose, J. 2021, Critical Appraisal of VAERS Pharmacovigilance: Is
the U.S. Vaccine Adverse Events Reporting System (VAERS) a Functioning Pharmacovigilance System? Science, Public Health Policy & the Law
Volume 3:100–129.
Hannah Ritchie, Edouard Mathieu, Lucas Rodés-Guirao, Cameron Appel, Charlie Giattino, Esteban Ortiz-Ospina, Joe Hasell, Bobbie Macdonald, Diana Beltekian and Max Roser (2020) - "Coronavirus Pandemic (COVID-19)". Published online at OurWorldInData.org. Retrieved from: 'https://ourworldindata.org/coronavirus' [Online Resource].
https://en.wikipedia.org/wiki/Intracellular_pH
Tetz G, Tetz V. Prion-like Domains in Spike Protein of SARS-CoV-2 Differ across Its Variants and Enable Changes in Affinity to ACE2. Microorganisms. 2022; 10(2):280. https://doi.org/10.3390/microorganisms10020280.
Sabaté R, Gallardo M, Estelrich J. An autocatalytic reaction as a model for the kinetics of the aggregation of beta-amyloid. Biopolymers. 2003;71(2):190-5. doi: 10.1002/bip.10441. PMID: 12767118.
Kauffman, S. A. (1986). Autocatalytic sets of proteins. Journal of Theoretical Biology, 119(1), 1–24. doi:10.1016/s0022-5193(86)80047-9.
Johansson J. Molecular determinants for amyloid fibril formation: lessons from lung surfactant protein C. Swiss Med Wkly. 2003 May 17;133(19-20):275-82. PMID: 12844270.
Wulf, MA., Senatore, A. & Aguzzi, A. The biological function of the cellular prion protein: an update. BMC Biol 15, 34 (2017). https://doi.org/10.1186/s12915-017-0375-5.
https://www.allergyresearchgroup.com/blog/enzymes-from-the-earth-lumbrokinase-nattokinase-and-heart-health/









My mother died of Alzheimer’s, my sister and first cousin died of ALS. One of many reasons I avoided the jab was the “prion-like spike proteins”, as they were described in early 2021. Now I know even more. Thank you !
Perhaps you should change your "stage" name to "Amazing Jessica" - definitely worth supporting...
I would need to re-read your articles a couple of times (as a non-doctor - of course) - thank you so much for your effort!
I wish I was as good in my job as you are in yours...