Integration of codon-optimized modified SARS-CoV-2 spike mRNA into human DNA
What we know and what we don't know yet...
Boy, the name of the genetic ‘filler’ in the lipid nanoparticles just keeps getting longer, doesn’t it?
A paper by Dhuli et al. has been doing the rounds entitled: “Presence of viral spike protein and vaccinal spike protein in the blood serum of patients with long-COVID syndrome”. It was published December 2023 in the European Review for Medical and Pharmacological Sciences by a group from Italy.1
This paper does suggest precisely what needs to be done en-masse yesterday: sampling and sequencing of DNA from various cell types of people - uninjected and injected, alike. BUT, and this is a strong but, this work does not prove integration. It needs reproduction and refinement.
N.B.: My intent is not to destroy this paper: on the contrary. Please read on. We need to get this right otherwise, they’ll dance us off into the ‘anti-vaxxer’ oblivion.
They sampled 81 people with “long-COVID” and claim to have found “viral spike protein” in one person following viral clearance, and “vaccine spike protein” in two people two months post injection. Let’s focus on the former. In the Supplementary Results they write:
PCR amplification using the specific primers targeting the BNT162b2 vaccine spike protein sequence resulted in nonspecific amplification in 38 out of the 80 samples.
Wow. That’s just shy of half of all of the samples. I think 80 is meant to be 81, by the way. Figure 1 shows a screenshot of the PCR products run on a gel to confirm the right sized products (amplicons). It is not clear to me why only 18 samples are shown as opposed to the 38 that resulted in ‘nonspecific amplification’, and also lanes 1, 6 and 11 (and 12) are questionable for presence of band. The band that showed up was at approximately 660 base pairs (bp).

So they were able to amplify something using the primers they selected that was approximately 660 bp. I’ll circle back to this.
One of the protocol issues I have with this work is that their ‘nested PCR’ seems to have been done with the same primers as for the regular PCR. For nested PCR, one needs a second set of primers to more precisely ‘pick-up’ and amplify the selected amplicon (the DNA amplified) to weed out the nonspecific bits that may have been amplified as a result of unexpected primer binding sites. In this case, they ran the PCR reaction products from 12 samples that showed strong bands twice more, using the same primer set as for the original PCR for these two subsequent rounds. Their second and third rounds of PCR yielded results that look like the following.
A single band of approximately 470 bp is clearly visible in all 12 samples, and in the case of 2 of the samples, 2 additional bands of approximately 660 bp were clearly visible. There are also some other smaller fragments that appear to be less than 300 bp.

In Figure S3 (Figure 3 below), they show the results from 13 samples (not sure why there are 13 - maybe one positive control?) following the third round of PCR. The strong single band of approximately 470 bp can been seen clearly.
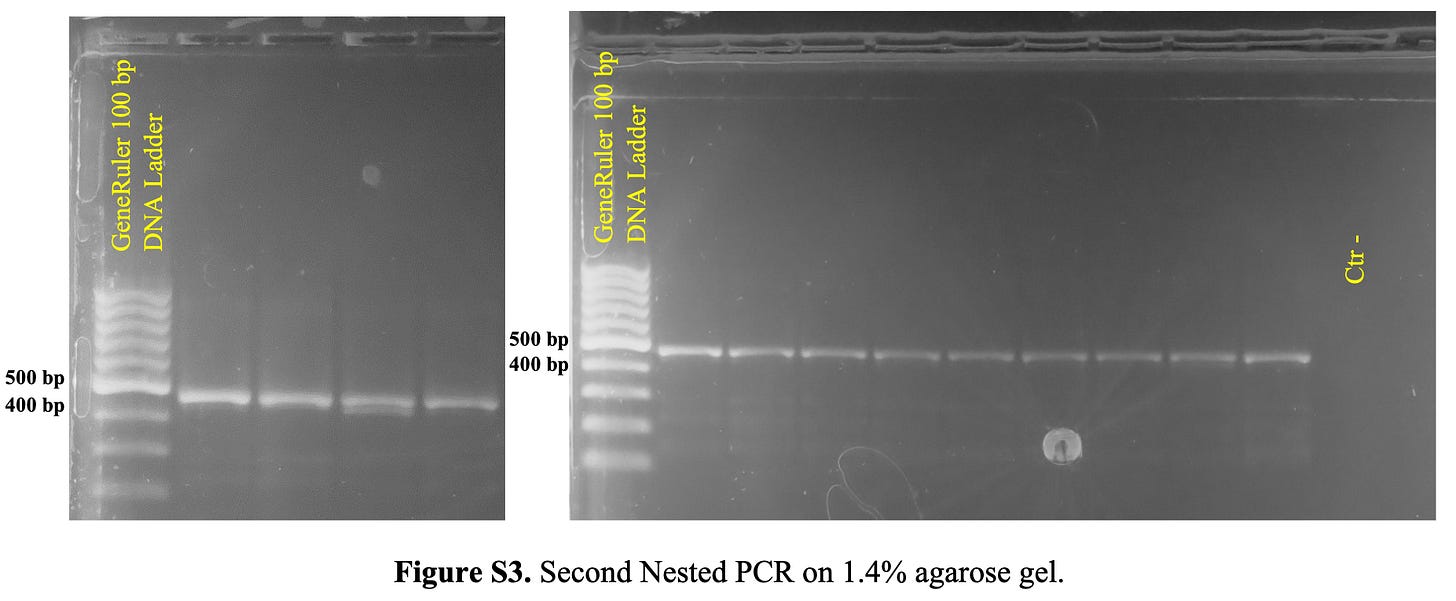
Based on the Pfizer sequence documented in the WHO International Nonproprietary Names Programme from September 2020 (you can download it from here), and the selected primers, the amplicon should be 400 bp. The forward primer as indicated in the Supplementary Results is CGAGGTGGCCAAGAATCTGA (red), and the reverse is TCTGGAACTAGCAGAGGTGG (blue). Don’t forget, they read from 5’ to 3’ so you need to reverse complement the reverse primer. In Figure 4, the selected primers from the study are shown in red for this sequence (BNT162b2 Pfizer/BioNTech) nestling the sequence to be amplified which sits at the end of the sequence.

The question for me becomes: why is the amplicon ~70 bp longer than the amplicon specified by the selected primers? I really like the consistency in bands of ~470 bp that they were able to produce - this is excellent, but what is that amplicon? My guess would be the appropriate amplicon consistent with the actual fragment that lies between the primers, which would mean that the WHO sequence does not match the sequence in the DNA samples. In this study, the “expected size of the fragment amplified by the BNT162b2” is not specified and neither is the original sequence representing BNT162b2. I will request this from the authors when I write to them.
Also, a huge guff I have with this particular result is the lack of description of and/or use of controls. Where is the positive control that would be the BNT162b2? Just RT-PCR that thing and run the products on a gel. Perfect 440 bp band should show up as positive control. What did they use a negative control? Was this water? Blood from an uninjected person?
Another protocol-related guff I have is the use of two kits to isolate the DNA: the E.Z.N.A.® Genomic DNA Isolation Kit, Omega Bio-Tek and the Exgene™ Blood SV kit. You can find this in their supplementary methods. The authors assessed “viral integration in patients’ leukocytes” which means they looked for DNA in white blood cells (leukocytes) originating from whole blood. The latter kit makes reference to this and cautions against having too few or too many leukoctyes: “The DNA yield from whole blood will depend on the number of white blood cells (WBCs, leukocytes) included in the sample.” The former kit can isolate mitochondrial DNA in addition to genomic and viral DNA. It is not a good idea to use 2 different kits for the same study because it adds potential confounders, and questions if things go wrong. Or right.
Apart from being put-off by the fact that they used 2 different kits, they only show 1 chromatogram that they claim “showed significant similarities between the detected spike protein fragments and the reference sequences of the SARS-CoV-2 spike protein (Figure S4)”.
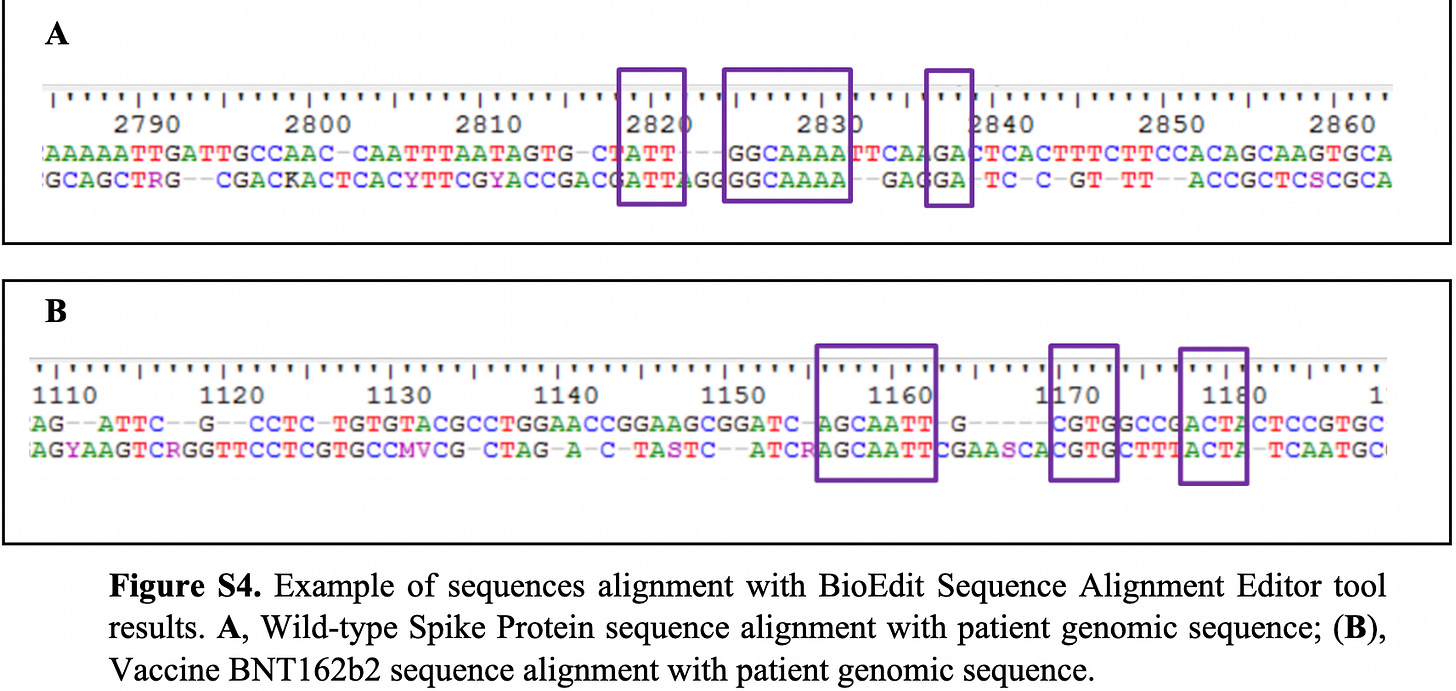
If I was comparing a DNA sequence sample from the leukocytes of an injected person with a BNT162b2 sequence sample, and my alignment looked like theirs as per Figure 5 (Figure S4), I would not feel that this was the evidence of integration that I might have been looking for. I wouldn’t call it nothing, but I would perform many, many more repeats prior to claiming integration. I would also show the full alignment for multiple comparisons.
Reminder: My intent is not to destroy this paper: on the contrary.
It is very valuable in its demonstration of protocol as per what we need to do post the billions-injected-DNA-contamination scandal. But I must read it as I would as if I was reviewing it: free from bias, and the fact is that I do have a number of concerns (as pointed out above) that require addressing. And yes, I am bringing these comments to the authors.
Perhaps more compelling evidence of integration is found in the paper referenced herein by Zhang et al. entitled: “Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues” published in PNAS in May, 2021.2 The authors demonstrate in vitro integration using human embryonic kidney 293 (HEK293) cells via the endogenous human retrotransposon LINE1 - a beautiful precursor to the Alden paper also clearly demonstrating reverse transcription of the modified mRNA from Pfizer BioNTeach BNT162b2 via LINE-1 in human liver cells.3
They write:
These results indicate that SARS-CoV-2 sequences can be integrated into the genomes of cultured human cells by a LINE1-mediated retroposition mechanism.
Ho ho ho. Merry Christmas.
The bottom line here is that we need to confirm these in vitro studies and do a whole lot of “in vivo” studies, ie: sample DNA from injected and uninjected people to get a good idea of whether or not - and how frequently - integration events are ensuing in people. My gut tells me this is happening, but we need strong and compelling evidence to eviscerate the compounding narrative of safety and efficacy of these modified mRNA products. It is simply malfeasant to make claims of non-integration since this has not been demonstrated. And I will repeat myself again, the onus in NOT ON US to show proof on integration - even though we are the ones generating the studies - it is on the manufacturers, the regulatory bodies, the governments, the 3-letter agencies - absolutely anybody who maintains the current CDC claim that “[The modified mRNA products] do not affect or interact with our DNA.”
Dhuli K, Medori MC, Micheletti C, Donato K, Fioretti F, Calzoni A, Praderio A, De Angelis MG, Arabia G, Cristoni S, Nodari S, Bertelli M. Presence of viral spike protein and vaccinal spike protein in the blood serum of patients with long-COVID syndrome. Eur Rev Med Pharmacol Sci. 2023 Dec;27(6 Suppl):13-19. doi: 10.26355/eurrev_202312_34685. PMID: 38112944
Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2105968118. doi: 10.1073/pnas.2105968118. PMID: 33958444; PMCID: PMC8166107
Aldén M, Olofsson Falla F, Yang D, et al. Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line. Curr Issues Mol Biol.. 2022;44(3):1115-1126. Published 2022 Feb 25. doi:10.3390/cimb44030073



Ho ho hmm 🤔
Thanks for your meticulous attention to detail, Jessica. This is how science is done. Let’s hope the authors respond to your valuable observations.
Oh, and merry Christmas Eve 🎄
"the onus in NOT ON US to show proof on integration - even though we are the ones generating the studies - it is on the manufacturers, the regulatory bodies, the governments, the 3-letter agencies - absolutely who maintain the current CDC claim that “[The modified mRNA products] do not affect or interact with our DNA.”"
Well, to at least design an experiment that we agree would verify this. I don't trust them to run it or report the results accurately.