We have evidence now that contaminating foreign DNA integration events are possible with respect to the COVID-19 modified mRNA products in a cancer cell line. You can read more about that here and here. Phillip Buckhaults is about to embark on a new project to test cancer cells from real people for such integration events. You can read about that here.
My daily posts summarizing science articles have not been as frequent because I am in a pensive phase after four years of going all-out. I need to stop and think more about what they actually did all this for, from a science point of view. Most of me thinks that this entire COVID thing was a ruse to provide a segue for the gene therapy era for which the modified mRNA LNP platform will be essential. You can listen to me talk about that here.
I want this article to be a review on the CRISPR/Cas system and its lab-made counterpart (the ‘gRNA-Cas-9 system’)1 , since I believe that this is going to be an essential component in the next stages of ‘personalized health care’, which I believe will be fully integrated into a ‘centralized health care system’ where data will be extracted and made available without restrictions between agencies, and where everyone’s genome will be known and perhaps even accessible and modifiable.
On CRISPR/Cas
The CRISPR/Cas system originates from bacteria archaea and is a system designed to cut specific pieces of DNA in foreign invaders (like bacteriophages) or plasmids - at specific places, as part of an ingenious immune defense system.2
We humans have absconded this genius system for use in gene therapy and genetic engineering. It is primarily used as a genome engineering tool to induce site-directed double-strand breaks in DNA to knock-out genes, for example. Cas-9 in particular, derived from Streptococcus pyogenes, is ‘a favorite’ with regard to genome editing tools.
On CRISPR - Clustered Regularly Interspaced Short Palindromic Repeats
CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e. anti-phage) defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.3

On Cas-9 - CRISPR-associated protein-9
Cas9 is a protein that plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome.4

On gRNA/sgRNA - (single) guide RNA
Guide RNA (gRNA) or single guide RNA (sgRNA) is a short sequence of RNA that functions as a guide for the Cas9-endonuclease or other Cas-proteins that cut the double-stranded DNA and thereby can be used for gene editing. In bacteria and archaea, gRNAs are a part of the CRISPR-Cas system that serves as an adaptive immune defense that protects the organism from viruses. Here the short gRNAs serve as detectors of foreign DNA and direct the Cas-enzymes that degrade the foreign nucleic acid.5
On PAM - Protospacer Adjacent Motif
A protospacer adjacent motif (PAM) is a 2–6-base pair DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system. The PAM is a component of the invading virus or plasmid, but is not found in the bacterial host genome and hence is not a component of the bacterial CRISPR locus. Cas9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence. PAM is an essential targeting component which distinguishes bacterial self from non-self DNA, thereby preventing the CRISPR locus from being targeted and destroyed by the CRISPR-associated nuclease.6
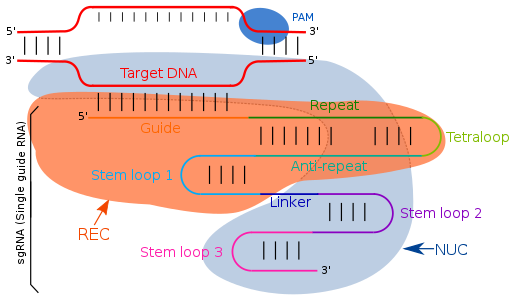
The amazing thing about this system, is that the CRISPR/Cas-9 complex can be simplified to a 2-component system to include the sgRNA and the Cas-9 protein, effectively creating an artificial Cas-9 system (gRNA-Cas-9 system). In doing so, this system can ‘find’ and cut any DNA target as specified by the gRNA. Do you guys understand the implications of this? It’s quite incredible. It means that the lab-made gRNA-Cas-9 system can be programmed to target any DNA sequence to enable modification of whatever gene you want. The other amazing thing about it is that is also ‘modular’: additional enzymes or modifications can be made to the system to enable not just double-stranded DNA breakage but nucleotide swap-outs7, activation (or silencing) of genes8 or gene therapy where DNA sequences are edited.9
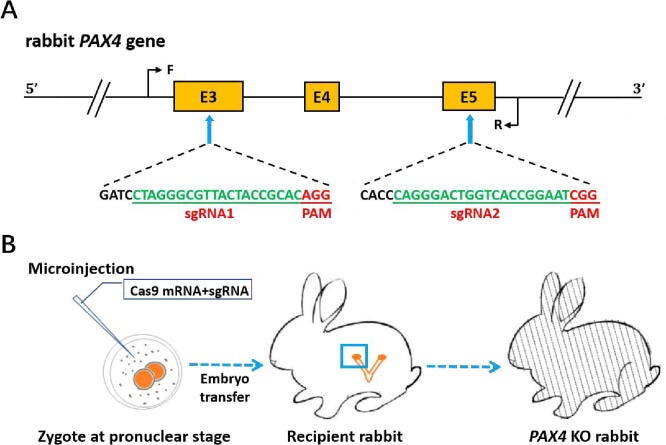
Nuclease domains nick DNA to knock-out certain genes. You can also deactivate the cutting domains, and fusing new enzymes, like deaminase to mutate specific bases, onto the Cas-9 protein. A study published in 2018 has demonstrated use of the CRISPR/Cas-9 system to knock out a gene (PAX4) linked to human diabetes mellitus in a rabbit model. Knock out the PAX4 gene, bunnies get sick.1011
Here’s a video that summarizes how we use CRISPR/Cas-9 system quite well.
Now that we all have a great understanding of the CRISPR/Cas-9 system and how humans are exploiting this system to do gene therapy and stuff, let’s get back to why we’re interested in this subject matter in the first place.
Recently, sequencing of COVID-19 modified mRNA Pfizer vials has revealed the presence of SV40 enhancer. Vials sequenced by Medical Genomics (work reproduced) revealed the enhancer as part of the plasmid map used for production of the modified mRNA - not disclosed by Pfizer or BioNTech in the ‘original plasmid map’ released - as indicated in this version of the Rolling Rapporteur report.12 Currently, both Health Canada (HC) and the European Medicines Agency (EMA) admit to its presence.13 This was just one bit of DNA discovered of the billions of fragments also discovered in both mono and bivalent Pfizer vials, by the way.
The SV40 enhancer is a very useful as a gene therapy tool because it’s a nuclear localization sequence (NLS): it’s very efficient at trafficking stuff to the nuclei of cells. You can read about that here.14 If this is the first time you’re finding out about this, then you’re probably wondering why in the ‘H’ this NLS is in this Pfizer plasmids/vials. It really doesn’t serve a function as per the manufacturing process, so it doesn’t need to be there. It’s a mammalian expression promoter.15 You would be right to wonder.
The concern that a few of us have expressed having discovered (and learned about) this superfluous DNA in the vials, is its potential integration into the genomes of the cells transfected by the modified mRNA LNPs, injected into billions of people as part of the COVID-19 injection roll-out madness (I wish Substack had an option for a creepy font). Random integration events can result in genomic instability and can lead to such things as cancer, especially if they occur in essential genes.

Evidence of integration events in human chromosomes 9 and 12 has indeed been recently reported by Medical Genomics’ Kevin McKernan. Having learned about PAMs here, I started thinking about the codon optimization aspect of the modified mRNA that increased the GC content. Remember, PAMs are 2-6 nucleotide fragments that allow Cas-9 to locate and interrogate DNA. “Specific interactions between Cas-9 and the PAM are required for effective binding of the target sequence and local strand separation prior to cleavage.”16 Oftentimes, they come in the form NGG, where N is any nucleotide.
An increase in CG content would theoretically increase the number of PAMs in the spike gene in the plasmid, wouldn’t it? If integration events of short DNA fragments are occurring, then the likelihood of a bit having a PAM-associated insert would be increased, would it not? If, horror of horrors, this was known prior, then pre-designed sgRNAs would be good-to-go. They would simply need to get to the nucleus and cut, cut, cut. This is entirely hypothetical. There is no need to believe this would even be logistically possible.
Somebody who goes by ‘M.A.’ uploaded a study entitled “The Pfizer Vaccine CRISPR Experiment” to the Zenodo preprint server that discusses the idea that the modified mRNA in the Comirnaty (and ‘he’ spells this wrong consistently as Cominarty) product, contains the coding material for CRISPR/Cas9.17 The reason I remember the spelling is because ‘Comir’ is part of the word onComir, as in: the microRNAs associated with cancer. In the name ‘Comirnaty’, miRNA itself is bookended between the ‘co’ and ‘ty’. Too bad it wasn’t a ‘zy’, eh? Would be a very ‘co-zy’ miRNA then.
Personally, I think this ‘person’ might be a fake since although they do have an ORCID ID, they do not have a name. And their email appears fake as well. I have sent a question to their listed email, but received no reply in days. It might be an agency psy-op because there’s no sign of Cas-9 in the coding material of the genes in the Pfizer plasmid discovered by Kevin McKernan. I checked. If it was there, it would be really obvious too because it’s big (1,018 bp).
I have two questions in my head. How readily would integration events with PAMs occur in any given transfected cell, and did anyone actually have this in mind? If a Cas-9 protein were to be instructed to be made as part of another ‘vaccine’, would the interaction with the Cas-9, the sgRNA (also introduced exogenously) and the integrated fragment result in cutting? Or is this about gene silencing or activation? Theoretically, you could introduce stop codons or alter a mutated gene using this method. You could also add transcriptional activators to the Cas-9 to activate transcription machinery of specific genes, ie: the modular aspect I mentioned previously. You could also inactivate transcription to physically block a gene. The possibilities are wide-ranging.
I am going to leave the following here and circle back to this at the end. This alignment is thanks to Dr. Ah Kahn Syed. Genius. And thanks for the tip on Ugene (it kicks MEGA’s ass for Mac OS), and the read through. N.B. I added a couple ‘control’ sequences - including the published WHO sequence: 11889 - to check for myself. It’s quite striking. Even in the Novavax product, there are 38 nucleotides ‘conserved/preserved’ among them all - not including the CGG which could constitute a PAM.

Questions to ponder: Why is this conserved fragment (1252-1289) in all of these products/vectors/sequences? Why are they sitting upstream from a PAM (CGG)? Could they be a target DNA for subsequent gRNA-Cas-9 system administration?
Pretty dark questions. What would possibly be the purpose, if the outcome of ‘installing’ this hardware in the genome - logistically - could never really be predictable or reproducible? Is it not predictable/reproducible?
An HIV gRNA-Cas-9 system tangent
I have a pretty wild imagination, and I can go pretty far in my imaginings of what might be possible with regard to science. Conventional science ‘knowledge’ as a backdrop can be … tricky, and I try not to be too stringent in my knowledge-base. What the hell do we know about anything, really? We don’t even know what consciousness is, but it ‘drives’ each and every one of us every day.
I can imagine the engineered Cas-9 system being used to cut the HIV genome out of the genomes of latently-infected human cells. And sure enough, people in the science community have been experimenting with this for many years.18 It’s a brilliant idea. It’s almost as if someone had the idea to put it there in the first place to see if they could get it out (kidding). I mean, our genome is ~8% retroviral (see Endogenous RetroVirus (ERV)), and these retroviral integrations are really important to our existence (we’ve evolved to co-opt certain retroviral proteins to serve in reproduction and development), but when you’re talking about something like HIV that eats CD4+ T cells for breakfast (and whose origin (in my opinion) are still ‘questionable’ (read the book ‘The River’)), you kind of don’t want that kind of virus integrating.
With regard to using the gRNA-Cas-9 system to get rid of HIV, you can also use it eat certain bits of the integrated HIV-1 genome to disable it. Take that, HIV. In a paper published in 2013, a codon-optimized gRNA-Cas-9 system was used to target the long terminal repeats (LTRs) of HIV-1. The designed gRNA was complementary to the LTRs. Using transfection, the gRNA along with a humanized Cas-9 was introduced into mammalian cells using plasmids.19 This idea is reminiscent of the trojan horse technology of the LNPs that, according to recent findings, also contain plasmids and plasmid DNA fragments.
The gRNA-Cas-9 system can also be used in the context of the ‘shock and kill’ strategy to eliminate HIV from the system. In this case, the gRNA-Cas9 system is used to activate latent HIV-1 viral reservoirs so that drugs (antiretroviral therapy (ART)) can be used to kill latently-infected cells. Kind of like smoking out the bees. A paper was published in 2022 entitled “SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8+ T-cells”, where the authors describe what sounds like an unexpected shock and kill result (strategy).20 Basically, the COVID-19 modified mRNA injections shook up (productively-engaged) HIV-Nef-specific CD8+ T cells - the ones that kill virally-infected cells - and resulted in a reduction in cell-associated HIV RNA, which is a measure of HIV persistence.
Smoke (latency-reversal agents)21 = modified mRNA COVID-19 shots; Bees = latent HIV; Beekeeper = antiretroviral treatment
The authors summarize their findings in the following:
Although this activity - incidental in the context of SARS-CoV-2 mRNA vaccines - was insufficient to drive measurable reductions in intact HIV DNA, there are multiple ways that future studies may intentionally optimize and leverage this built-in latency reversing agent (LRA) activity to enable reservoir reductions by mRNA vaccines encoding HIV antigens.
Translation: Let’s use the modified mRNA tech as an LRA in the future - ie: advance the platform and spill it over to the HIV setting. The possibilities are endless!
There has been a lot of HIV-related ‘stuff’ in the woodworks with regard to the COVID-19 saga, and it has really left me wondering why. And how. Besides the fact that the spike protein is structurally and compositionally similar to gp120, why was there a specific test group in clinical trials for people who were deemed HIV positive, and why were people being encouraged to be screened for HIV post injection?22 It has been reported that other vaccines can induce increases in cell-associated HIV RNA, and it has also been reported that increases in cell-associated HIV RNA can be accompanied by modest and transient increases in HIV-p24-specific CD8+ T-cell responses, so does this explain why?2324 Were certain people in these fields just curious to know the effects of this new, experimental technology on HIV? Maybe.
To me, there seems to be a lot of ‘surprising’ overlap. I mean, yeah, regular screening was probably reduced because of the psychotic 2-weeks to fatten the wallets thing, but still. The aforementioned paper actually states that the “rate of acute HIV infection (AHI) diagnoses per day was significantly higher during the pandemic compared with the prior 4 years”. I personally don’t buy that this is due to higher frequency testing exactly because routine HIV screening in health care settings was reduced (these likely would have balanced each other out, if anything, in my opinion), so what’s with the higher frequency HIV diagnoses? We’re talking about acute infection - the beginning; new infections - so, what is going on here? Maybe more people were being promiscuous? More IV drug use?
It is unlikely that the p24 antigen as per HIV antigen test would have anything to do with SARS-2 presence, so as far as SARS-2 making an HIV test ‘come up positive’, I doubt this would ever happen. So what we appear to be facing is a real uptick in people with acute HIV infections. But why?
In my research for this article, I also discovered something that I hadn’t previously known. HIV is selective about where it integrates into the human genome. In fact, it really likes chromosome 11.25 It’s also been shown that HIV integration disproportionately targets cancer genes.26 My thinking, based on logic, tells me that integration events would occur in places where the DNA was active, ie: uncoiled/accessible. I have to repost this amazing animation because it really nicely demonstrates visually how, and at what point, the unzipping of the DNA occurs (4:09), for example, during transcription.
Why am I talking about this? Well, because if someone wanted to use the CRISPR/Cas-9 system to activate a gene, they would have a better time of it if they knew where the gene was.
The reason I went into this HIV story is because I wanted to demonstrate the proof of concept (in vivo) use of the engineered gRNA-Cas-9 system in the context of HIV - a real-life retrovirus that is problematic for humans - that currently has no sustainable solution. There’s also this, but never mind. The reason I needed to investigate how far along they are (or at least what has been published), is because I wanted to find out how easy it would be to actually deploy the engineered gRNA-Cas-9 system using say, LNPs as a delivery system of the gRNAs and humanized Cas-9 using plasmids. And if it was attainable as a research goal, would it be conceivable that anyone would allow this to go beyond research?
The next step in this investigation is to look for Cas-9 and the gRNA complementary to this:
GAT CGC CGA CTA CAA CTA CAA GCT GCC CGA CGA CTT CAC CGG
To be continued…
Xu Y, Li Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput Struct Biotechnol J. 2020 Sep 8;18:2401-2415. doi: 10.1016/j.csbj.2020.08.031. PMID: 33005303; PMCID: PMC7508700
Barrangou R (2015). "The roles of CRISPR-Cas systems in adaptive immunity and beyond". Current Opinion in Immunology. 32: 36–41. doi:10.1016/j.coi.2014.12.008
https://en.wikipedia.org/wiki/CRISPR
https://en.wikipedia.org/wiki/Cas9
https://en.wikipedia.org/wiki/Guide_RNA
https://en.wikipedia.org/wiki/Protospacer_adjacent_motif
Fortunato F, Rossi R, Falzarano MS, Ferlini A (2021-02-17). "Innovative Therapeutic Approaches for Duchenne Muscular Dystrophy". Journal of Clinical Medicine. 10 (4): 820. doi:10.3390/jcm10040820
Jiang F, Doudna JA (2017-05-22). "CRISPR–Cas9 Structures and Mechanisms". Annual Review of Biophysics. 46 (1): 505–529. doi:10.1146/annurev-biophys-062215-010822
Demirci S, Leonard A, Haro-Mora JJ, Uchida N, Tisdale JF (2019), "CRISPR/Cas9 for Sickle Cell Disease: Applications, Future Possibilities, and Challenges", Cell Biology and Translational Medicine, Volume 5, Advances in Experimental Medicine and Biology, vol. 1144, Cham: Springer International Publishing, pp. 37–52, doi:10.1007/5584_2018_331
Yuanyuan Xu, Yong Wang, Yuning Song, Jichao Deng, Mao Chen, Hongsheng Ouyang, Liangxue Lai, Zhanjun Li, Generation and Phenotype Identification of PAX4 Gene Knockout Rabbit by CRISPR/Cas9 System, G3 Genes|Genomes|Genetics, Volume 8, Issue 8, 1 August 2018, Pages 2833–2840, https://doi.org/10.1534/g3.118.300448
Xu Y, Li Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput Struct Biotechnol J. 2020 Sep 8;18:2401-2415. doi: 10.1016/j.csbj.2020.08.031. PMID: 33005303; PMCID: PMC7508700
Rapporteur Rolling Review critical assessment report. Quality aspects. COVID-19 mRNA Vaccine BioNTech. Modified mRNA, encoding full length SARS-CoV-2 spike protein. EMEA/H/C/005735/RR/xxx
Speicher, D. J., Rose, J., Gutschi, L. M., Wiseman, D. M., PhD, & McKernan, K. (2023, October 19). DNA fragments detected in monovalent and bivalent Pfizer/BioNTech and Moderna modRNA COVID-19 vaccines from Ontario, Canada: Exploratory dose response relationship with serious adverse events. https://doi.org/10.31219/osf.io/mjc97
Young JL, Benoit JN, Dean DA. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003 Aug;10(17):1465-70. doi: 10.1038/sj.gt.3302021. PMID: 12900761; PMCID: PMC4150867
https://blog.addgene.org/plasmids-101-the-promoter-region
Konstantakos V, Nentidis A, Krithara A, Paliouras G. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning. Nucleic Acids Res. 2022 Apr 22;50(7):3616-3637. doi: 10.1093/nar/gkac192. PMID: 35349718; PMCID: PMC9023298
M.A. (2022). The Pfizer Vaccine CRISPR Experiment. https://doi.org/10.5281/zenodo.6210570
Xiao Q, Guo D, Chen S. Application of CRISPR/Cas9-Based Gene Editing in HIV-1/AIDS Therapy. Front Cell Infect Microbiol. 2019 Mar 22;9:69. doi: 10.3389/fcimb.2019.00069. PMID: 30968001; PMCID: PMC6439341
Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. PMID: 23974631; PMCID: PMC3752613
Stevenson, E.M., Terry, S., Copertino, D. et al. SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8+ T-cells. Nat Commun 13, 4888 (2022). https://doi.org/10.1038/s41467-022-32376-z
Spivak AM, Planelles V. Novel Latency Reversal Agents for HIV-1 Cure. Annu Rev Med. 2018 Jan 29;69:421-436. doi: 10.1146/annurev-med-052716-031710. Epub 2017 Nov 3. PMID: 29099677; PMCID: PMC5892446
Stanford KA, McNulty MC, Schmitt JR, et al. Incorporating HIV Screening With COVID-19 Testing in an Urban Emergency Department During the Pandemic. JAMA Intern Med. 2021;181(7):1001–1003. doi:10.1001/jamainternmed.2021.0839
Stevenson, E.M., Terry, S., Copertino, D. et al. SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8+ T-cells. Nat Commun 13, 4888 (2022). https://doi.org/10.1038/s41467-022-32376-z
Yek, C. et al. Standard vaccines increase HIV-1 transcription during antiretroviral therapy. AIDS 30, 2289–2298 (2016)
Schröder, Astrid R.W. et al.. HIV-1 Integration in the Human Genome Favors Active Genes and Local Hotspots. Cell, Volume 110, Issue 4, 521 - 529
Singh PK, Plumb MR, Ferris AL, Iben JR, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH, Kvaratskhelia M, Levin HL. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015 Nov 1;29(21):2287-97. doi: 10.1101/gad.267609.115. PMID: 26545813; PMCID: PMC4647561



If I understood all of this it would blow my mind. Fortunately, a sufficient degree of ignorance combined with a certain intellectual laziness to dig deeper into all of this stuff curtails my wildest imaginings.
So nobody 'invented' CRISPR; some humans noticed that archaeons and bacteria used it as a neat trick to protect themselves from infection and so decided to exploit it, patent it and make a load of money in the process, as well as perhaps deciding that they would appoint themselves as the 'accelerant' of human evolution, thus deposing God and Charles Darwin.
AFAIK, my genome hasn't been harvested yet by these psychos for their Universal Database because I refrained from being forced to or voluntarily submitting to shoving an ethylene soaked extra long cotton bud up my nose halfway to my brain in order to test for the presence of a virus which was not causing the presence of any symptoms. I intend to try to keep it that way until death do us part, on account of my genome being a very Selfish Genome. I don't mind God or Darwin opening it up to take a looksie now and then, but I draw the line at psychos loaded with cash generated from patents doing so.
I wonder how inherited genomic integration might effect the next generation?