CRISPR-Cas9 modifications to exploit the production of biomolecules from bacteria
The hits just keep on comin'!
Yet another Nature article published on January 21, 2025 entitled: “Engineered CRISPR-Cas9 for Streptomyces sp. genome editing to improve specialized metabolite production”1 is on my radar. Basically, any use of CRISPR-Cas9 is in my radar: I want to know when and how it’s being used.
Now by now readers already know the obvious right? If you need a refresher on CRISPR-Cas9, please head here.
Have we been CRISPRed for the coming Cas-9?
We have evidence now that contaminating foreign DNA integration events are possible with respect to the COVID-19 modified mRNA products in a cancer cell line. You can read more about that here and here. Phillip Buckhaults is about to embark on a new project to test cancer cells from real people for such integration events. You can read about that
And if you need a refresher on gene editing/swapping/making a GMO, please head here.
A new Nature paper describing a rub-on "vaccine"
A paper hit the presses on December 11, 2024 entitled: “Discovery and engineering of the antibody response to a prominent skin commensal”. You can read the Coles Notes written by Bruce Goldman of Stanford Medicine News Center here. Thanks to Anand Ram for bringing this paper to my attention.
There is a bacterial species called Streptomyces that makes all sorts of nifty biomolecules like anti-cancer agents and antibiotics, aka: secondary metabolites. Like so many things in nature, we exploit these bacteria in specific ways to use these biomolecules for study and therapeutics. But we always seem to want more.
Among many things, we use something called biosynthetic gene cluster (BGC) refactoring - a type of genetic engineering - to enhance gene expression to increase production of these precursor metabolites in Streptomyces species.
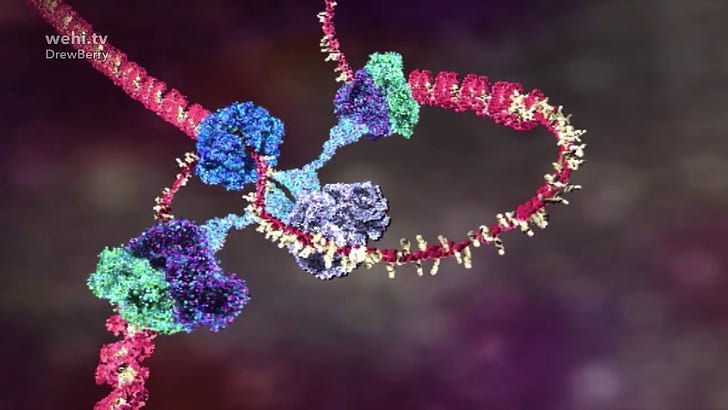
Biosynthetic gene clusters (BGCs) are groups of genes that work together to produce specialized metabolites, such as antibiotics and other secondary metabolites in microorganisms like bacteria. Refactoring these clusters involves redesigning or modifying the genetic architecture of BGCs to enhance, alter, or study the production of these compounds. We use the CRISPR-Cas9 system for precise genome editing, which can be utilized in refactoring to make targeted changes.
One of the commonly-used Cas9s used for such editing comes from Streptococcus pyogenes. Apparently, however, genetic engineering using CRISPR-Cas9 of Streptomyces in this way is hard because of the high guanine and cytosine (GC) content in its genome. One of the reasons it is hard is because the commonly-used Cas9 from Streptococcus pyogenes recognizes the ‘-NGG’ as a protospacer adjacent motif (PAM) sequence. The CRISPR-Cas9 system can also cleave at sites with sequences similar to the target (called off-target cleavage), and this produces undesired results: no secondary metabolites, and it’s all because of those plentiful Cs and Gs.
The authors got around this difficulty by modifying Cas9 from S. pyogenes. In really layman’s terms, they added “D”s (aspartate) to the end or beginning of the Cas9 sequence using a glycine-serine (GS) linker (aka: Cas9-BD).
With reduced toxicity when expressed in the strain, Cas9-BD was employed for genome editing of Streptomyces sp. to enhance secondary metabolite production in various ways.
The modified proteins exhibited low toxicity to Streptomyces when expressed at high levels.
Using Cas9-BD the production of secondary metabolites in Streptomyces was dramatically improved in a single experiment using a library containing multiple sgRNAs.
Without going into any more detail, the authors engineered the genome of Streptomyces using a modified S. pyogenes Cas9 to enhance gene expression to increase production of these precursor metabolites in Streptomyces species.
That’s it for this article.
I think CRISPR-Cas9 is here to stay folks.
Kim, D.G., Gu, B., Cha, Y. et al. Engineered CRISPR-Cas9 for Streptomyces sp. genome editing to improve specialized metabolite production. Nat Commun 16, 874 (2025). https://doi.org/10.1038/s41467-025-56278-y




I’m sorry about repeating this comment elsewhere, but I was having trouble receiving my emails and I just wanted to get this news out: Becerra is still in charge at HHS and they just awarded Moderna millions of dollars to accelerate development of mRNA bird flu “vaccine”! Get this guy out of there! End the very dangerous mRNA “vaccines” now!
Yes Dr. Jessica, I believe you’re right . It’s not about saving lives , it’s about the money to be made . 💰