A recent preprint links SV40 enhancer to cancer but more importantly, to AID and somatic hypermutation
Is the generation of antibody diversity on the line?
In January 2024, a preprint was uploaded to the bioRxiv preprint server entitled: “The SV40 virus enhancer functions as a somatic hypermutation targeting element with potential oncogenic activity”.
In a Venice.ai-summarized nutshell, the paper established the following:
The SV40 enhancer has been found to function as a somatic hypermutation-targeting element with potential oncogenic activity. This was discovered through research on the Merkel cell polyomavirus (MCPyV), which causes Merkel cell carcinoma in humans by expressing truncated large tumor antigen (LT) due to APOBEC cytidine deaminase family enzymes induced mutations. Activation-induced cytidine deaminase (AID), a member of the APOBEC family, initiates the antibody diversification process known as somatic hypermutation (SHM). The SV40 enhancer's ability to target AID and induce SHM has implications for the development of cancer and the immune system's response to pathogens.
Kevin McKernan has written an article outlining the link to cancer due to the presence of SV40 (in droves) in the modified RNA COVID-19 injectable products (Pfizer). You can read that here.
Here is the phrase in the abstract of the preprint that I would like to focus on.
We demonstrate that the SV40 enhancer has strong [somatic hypermutation] (SHM) targeting activity in several cell types.
Now this is no easy subject matter to grasp - even for an immunologist - but I will do my best to describe it so that you all can understand. If you’re well-versed in reading science articles, read this one called “AID and somatic hypermutation”.1
One of the silver linings of the COVID scam, is that most of you probably know what antibodies are because of it. For those who do not, antibodies are secreted immunoglobulin molecules derived from the membrane-bound immunoglobulin molecules on a B cell. The process by which these antigen-specific secreted proteins are produced is quite complex. It starts in the bone marrow, and ends up in the blood (plasma), tissue fluid, tears (mainly IgA), saliva, breast milk (colostrum, specifically) and/or lymph nodes.
Antibodies serve many functions - from complement activation to binding and removal of pathogens - and come in five major isotypes (or classes) including the now infamous IgG isotype - infamous because of the IgG4 subclass hoopla. One of the most important and shared functional aspects of antibodies is their ability to bind their cognate antigens as tightly as possible, ie; with high affinity.
So what we’re going to get into in this article is how this tightness is established and what the implications of messing with that might be. The process of tightening the bond is called affinity maturation and it involves SHM combined with antigen-exposure via antigen-presenting cells to promote competitive selection of the ‘best’ B cell receptors (BCRs): only the B cells with the highest affinity BCRs will survive to become top notch potential antibody factories.
Thus, from a diverse repertoire of BCRs on B cells that arise from SHM, emerge highly specific receptors that will end up being the templates for secreted antibodies - trained on antigen, and highly specific.
Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes.2
We can’t fully understand the concept of SHM until we break down the component parts of the immunoglobulin molecule and how its genes are subject to mutation to allow for the generation of antibody diversity. We also can’t talk about SHM without knowing more about B cells.
On B cells
B cells function in what we call the humoral immunity component of the adaptive immune system - an essential branch of immunity. It is adaptive in that it can adapt to, respond to and remove new antigens that the body meets, like new viruses. They are actually called B cells because they were first discovered to mature in the Bursa of Fabricius found in the cloaca of birds. We humans don’t have this organ; our B cells mature in the bone marrow. Bakaw.
Our lymphatic system is quite systemic as you can see. B cells originate in the bone marrow (a primary lymphoid organ) from pluripotent stem cells, specifically from hematopoietic stem cells (HSCs) that differentiate into lymphoid progenitors and in the case of B cells, pro-B and pre-B cells. As they mature, developing B cells move toward the center of the marrow, and even though most B cells are thought to mature in the spleen, some B cells do stay in the marrow to fully mature. These mature B cells have undergone a selection process such that their BCR is not auto-reactive; that is: autoimmunity won’t be a problem. Mature B cells become activated upon encountering antigen - their BCR will recognize an antigen and bind to it. They will then migrate to secondary lymphoid organs (spleen and lymph nodes), where they undergo further differentiation and maturation, ultimately giving rise to antibody-secreting effector cells.

Try to visualize the lives of B cells as they physically change location and phenotype as they mature, differentiate and proliferate. The life of a B cell will be complete with the formation of long-lived memory cell pools and/or plasma B cells that pump out high affinity antibodies that arise as a by-product of SHM.
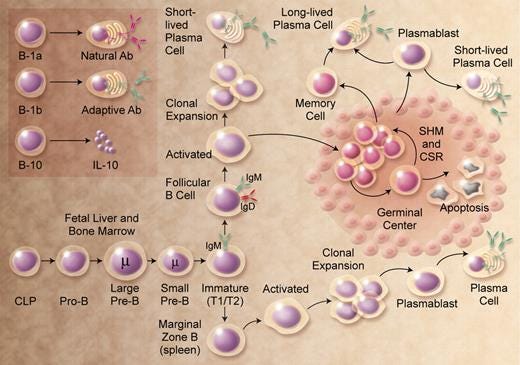
SHM occurs in the germinal centers of lymph nodes. Germinal centers are the places in the lymph nodes where the ‘seeds of the B cells’ grow. There, they multiply, differentiate and undergo SHM. By the way, the ‘somatic’ part of SHM refers to an individual’s immune cells (not germline cells) that divide by mitosis. The ‘hypermutation’ part of SHM refers to an elevated rate of spontaneous mutations in a cell’s DNA: the immunoglobulin gene segments are the sites of hypermutation.
Figure 4 below will help you visualize the process that B cells go through - from naïve to either memory or plasma cells - complete with SHM along the way.
Naïve B cells have not been exposed to antigen yet, whereas memory B cells have been; they have been primed to recognize a specific antigen having already met antigen. Naïve B cells can be found hanging out in the tonsils, spleen and lymph nodes, and memory B cells circulate around the body - for up to decades - looking for their respective cognate ligands on pathogens.
Whether naïve or memory type, when a B cell is ‘activated’ by an antigen, it can proliferate and differentiates into an antibody-secreting effector cell, known as a plasmablast or plasma cell. It is during proliferation that the B-cell receptor locus (the specific, fixed position on chromosome) undergoes an extremely high rate of somatic mutation - way higher than normal mutation rates across genome, as shown below. Otherwise, when activated, its destiny can be a memory cell.
Let’s get deeper on immunoglobulin molecules to get a better handle on this mutation thing. We’re gettin’ there.
The immunoglobulin molecule (as shown below) is made up of what we call constant regions (the C parts), and variable regions (the V parts). The ‘L’s refer to light chains: 25 kDa in size, and the ‘H’s refer to heavy chains: 50 kDa in size. As shown in the schematic below of IgG structure, there are 4 parts comprising 3 constant regions, and 1 variable region at the ‘Y’ end of the molecule on the heavy chain, and 2 parts comprising 1 constant region and 1 variable region on the light chain. SHM occurs in the variable regions of both the heavy and light chains. It is amazing that as part of antibody isotype switching from say, IgM to IgG, the constant region of the heavy chain is changed, but the variable region of the heavy chain is not. It is in this way that the function of the antibody can change, but not the specificity for antigen. Isn’t that neat?!

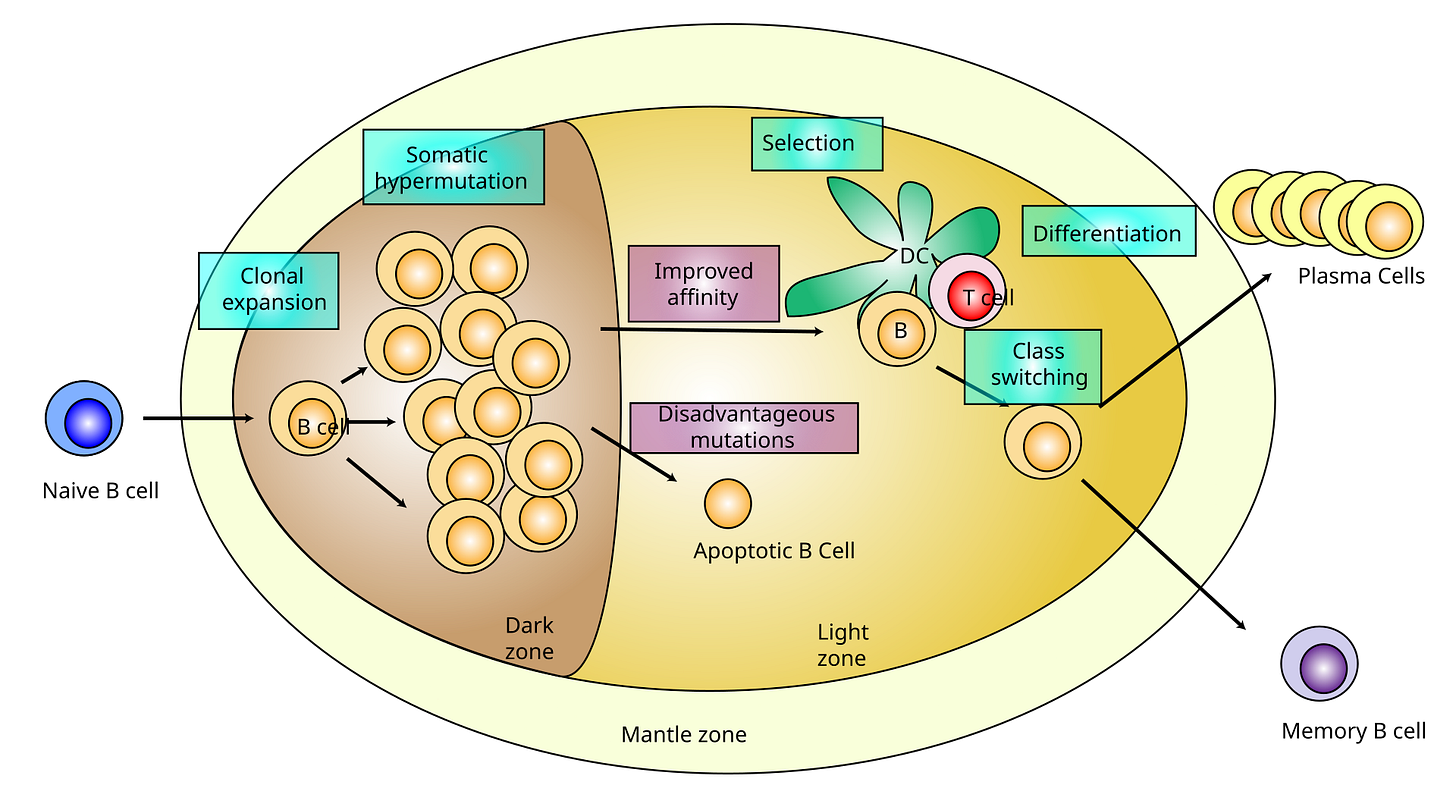
So the tip of the antibody structure where you see the V marked is where you get variability happening, and the hypervariable regions marked with white lines is where SHM occurs (Figure 5). Figure 6 shows another representation of where SHM can occur.

So how exactly does this affinity maturation/SHM go down?
The mutations that we’re talking about occur mostly at concentrated hypervariable regions called "hotspots" in the DNA that correspond to the sites involved in antigen recognition on the immunoglobulin molecule: that bit at the end of the ‘Y’ part of the molecule. The "hotspots" of somatic hypermutation vary depending on the base that is being mutated, and it is this ‘directed’ hypermutation that allows for the selection of B cells that express immunoglobulin receptors that possess an enhanced ability to recognize and bind specific foreign antigens.
Through sequencing data, it was found that there are specific motifs associated with targeted mutation. These motifs are enriched in the variable regions and thus the mutations are more pronounced in these variable gene sequences.
Watch this video. It’ll help.
Now here’s the clincher of this article. If you watched the wideo, you will have learned that a key enzyme necessary for somatic hypermutation to occur is called activation-induced cytidine deaminase (AID): the so-called MASTER CATALYST. It converts cytosine into uridine through something called a deamination process. Deamination just means the removal of an amino group from a molecule, like ammonia in the case of cytosine deamination to uracil.
When this deamination happens to a cytosine, there ends up being a mismatch of G with U, instead of G with C. Remember our base-pairing rules? C-G and A-T, right? Even though the mutation is from a C to a U, the cell's DNA replication machinery recognizes the U as a T (remember DNA uses Thymine (A, G, C, T) and RNA uses Uracil (A, G, C, U).
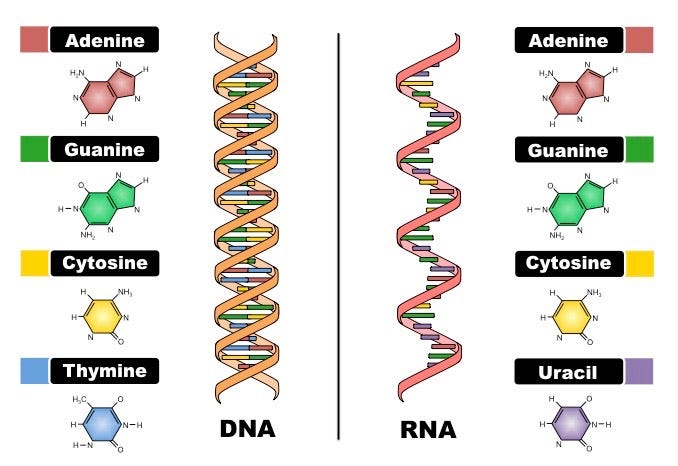
This means that the original C:G base pair is mutated to a U:G, ‘seen’ as a T:G and converted to a T:A base pair. These mutations induced by AID don’t just happen anywhere in the DNA. Remember the hotspots? Thanks to those sequencing studies I mentioned34, we know that there are specific motifs that the AID likes to target for mutation. These motifs are RGYW (i.e. A/G G C/T A/T) for a G, WRCY for a C, WA for an A and TW for a T.5 If the AID ‘sees’ an RGYW sequence, for example, mutations are likely. Which means a new BCR.
The mutations that take place are all pretty random and involve high fidelity DNA repair enzymes, combined with error-prone DNA polymerases. Since these mutations occur in the context of rapidly proliferating B cell populations as indicated above in Figure 4, the end product are thousands of B cells, all with slightly different BCRs with varying antigen specificity. B cells in germinal centers tend to divide rapidly and frequently having been activated. For the purposes of strong binding to antigens, the immune system selects for the mutations that lead to highest affinity (the strength and duration of the interaction between the B cell and the antigen) binding of BCRs and antigens. This is called positive selection. So it boils down to out-competition of B cells with the highest affinity binding BCRs whereby ultimately, the B cells with the highest affinity BCR will subsequently differentiate to plasma cells to produce high affinity antibodies.
The key part of this mechanism of SHM that deserves attention for the purposes of this article is the AID: this enzyme that changes Gs to Us.
I hope to this point my dear readers have a decent conceptualization of the B cell’s life journey and how important SHM is to BCR diversity. Brace yourselves, we’re moving on.
Now here’s the scary part that the preprint addresses. SV40 enhancer, the very same one found in the vials sequenced by Kevin McKernan, the very same one acknowledged to be present as ‘residual DNA fragment[s]’ by Health Canada (HC), the FDA and the European Medicines Agency (EMA), targets SHM machinery, specifically AID which of course, is responsible for initiating SHM.
Here are some possible suggested outcomes of introducing SV40 to the SHM environment:
Altered Antibody Affinity and Specificity: SV40’s potential to target AID could lead to aberrant mutations in immunoglobulin genes, resulting in altered antibody affinity and specificity. This might impact the effectiveness of the immune response.
Increased Oncogenic Activity: The SV40 virus enhancer’s ability to target AID could introduce oncogenic mutations, potentially contributing to tumorigenesis. This is supported by the observation that SV40 is associated with several human cancers, including mesotheliomas, brain and bone tumors, and lymphomas.
Error-Prone Repair Mechanisms: The introduction of SV40 could activate error-prone repair mechanisms, leading to an increased frequency of insertions, deletions, and point mutations in the vicinity of DNA breaks. This might result in the creation of novel oncogenic fusion genes or the disruption of tumor suppressor genes.
Aberrant Expression of APOBEC Enzymes: SV40’s interaction with SHM elements could lead to aberrant expression and targeting of APOBEC cytidine deaminase family enzymes, which are known to induce mutations in viral genomes. This might result in the creation of novel oncogenic mutations or the selection of pre-existing mutations.
Disruption of Normal SHM Process: The introduction of SV40 could disrupt the normal SHM process, potentially leading to impaired antibody diversification and reduced immune responses.
This last point was the one that concerned me the most. If we have impaired antibody responses, we have a broken immune system.
I am going to leave this article at this for now. We need to shout the DNA story from the mountain tops. This story is gaining incredibly important momentum in Australia right now thanks to the hard work and diligence of some exceptional and intrepid men and women. You can read about that here.
Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159-91. doi: 10.1016/S0065-2776(10)05006-6. PMID: 20510733; PMCID: PMC2954419
https://en.wikipedia.org/wiki/Somatic_hypermutation
Dunn-Walters, DK; Dogan, A; Boursier, L; MacDonald, CM; Spencer, J (1998). "Base-specific sequences that bias somatic hypermutation deduced by analysis of out of frame genes". J. Immunol. 160: 2360–64. doi:10.4049/jimmunol.160.5.2360
Spencer, J; Dunn-Walters, DK (2005). "Hypermutation at A-T base pairs: The A nucleotide replacement spectrum is affected by adjacent nucleotides and there is no reverse complementarity of sequences around A and T nucleotides". J. Immunol. 175 (8): 5170–77. doi:10.4049/jimmunol.175.8.5170
https://www.bioinformatics.org/sms/iupac.html - thanks Kevin!




The SV40 was known to promote cancer. It was not needed to get your cells to make spike protein to produce antibodies. There is no reason for SV40 to be in these jabs and it had to be put there deliberately. So the jabs were designed either to reduce the population or to boost sales of Pfizer's cancer drugs. And the fact that Pfizer hid the presence of SV40 is very damning. Heads must roll.
Thank u and enjoyed your talk with Steve Kirsch last night . I really believe the tumor surveillance system that Kevin M discusses wipes out the mitochondria within that system . These evil scientists went after the powerhouse .