The EMA leaks demonstrated lots of consternation from the regulators about the dsRNA levels in the vaccines. We only have EMA leaks for Pfizer so its unclear to what extent this is occurring in Moderna but Moderna’s publication records and patent publications imply it is very much on their radar.
Moderna and Pfizer have published at least two papers focused on methods to reduce the dsRNA content in their vaccines.
Moderna Engineered a mutated form of T7 polymerase to reduce dsRNA formation in 2023 (Dousis et al)
1)Dousis- https://www.nature.com/articles/s41587-022-01525-6
BioNtech published a paper optimizing mono, di and trinucleotide ratios to provide simultaneous capping (tri nucleotides) with modRNA synthesis (Ziegenhals et al).
2)Ziegenhals- https://www.frontiersin.org/articles/10.3389/fmolb.2023.1291045/full
Patent_Sun and Maria Gutschi had an interesting thread on some Moderna patents that imply the dsRNA and dsDNA contamination may be inversely correlated. Less dsDNA = more dsRNA and vice versa.
Why am I in this rabbit hole?
In the cannabis field, we need better tools to hunt for dsRNA viruses that impact plant yield. We have great tools in place to survey the cannabis genome, fungal and bacterial microbiomes and the dsDNA viral genomes with one simple assay known as Whole Genome Sequencing.
But this dsDNA based method is missing 80% of the pathogenic viruses which are RNA viruses and viroids.
Sequencing RNA can be expensive as the vast majority of the RNA in a plant sample is rRNA, tRNAs and host ssRNA gene expression. Ideally, one would pull out only the dsRNA and sequence just those molecules to improve efficiency, sensitivity and viral discovery as described by Javaran et al below.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7447037/
So one can use various RNases to remove ssRNA and dsDNA and only sequence dsRNA. This would be a helpful assay for us to have in place to screen for dsRNA viral genomes in cannabis plants.
But to iron this assay out, we need some high quality ssRNA contaminated with dsDNA to ensure the enzyme cocktails we have are effective at removing this background.
So lets make a mock blend as a control. Mix in dsRNA like circular rod-shaped Hop Latent Viroid (HpLVd) with the ‘ssRNA’ from the vaccines and see if we design a enzyme cocktail that will eliminate the dsDNA, ssRNA from the vaccines while leaving the HpLVd genome alone. HpLVd isn’t perfectly doublestranded RNA (see the bubbles or stem loops in molecule below) so we may need a few other models templates but we have this one laying around.
What about the vaccines? Are they a perfect model for this? Are they pure ssRNA or do they have any dsRNA?
They are supposed to be heavily regulated to have very little dsRNA (as it triggers Toll Like receptors and RNA interference pathways) but as Maria Gutschi and Patent_Sun have pointed out, this appears to be another regulation they were given a hall pass on.
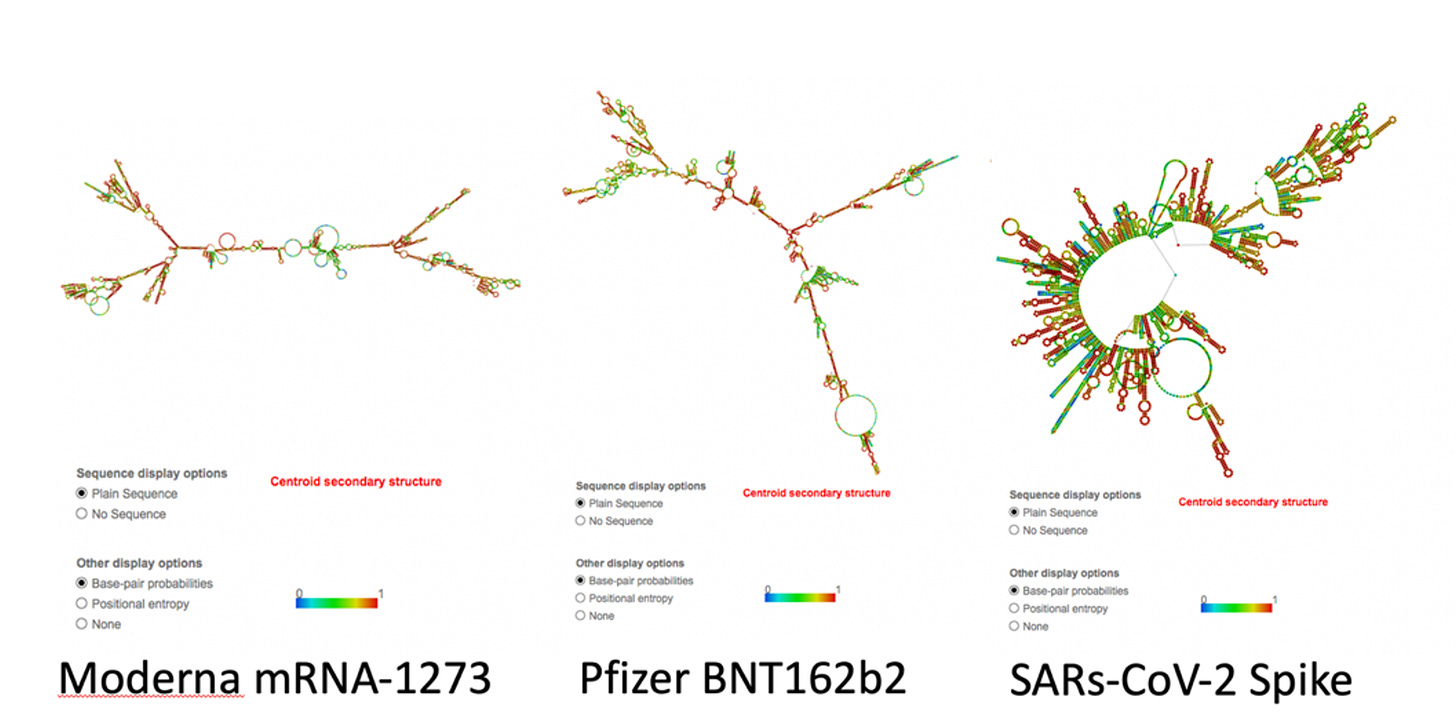
We anticipated a lot of dsRNA once we saw that their codon optimization massively shifted the GC content of the mRNAs. Once the GC content increases, the ssRNA becomes much more ‘sticky’ and binds to itself. Parr et al have also shown the addition of N1-Methyl-PseudoU will radically increase the Melting temperature of RNA and also increase its ‘stickyness’ and propensity to form dsRNA. See Figure 2 in Parr et al.
You can see the impact this GC shift has had on the structure of the RNA below (RNAfold). Many more stretches of dsRNA. This means the codon optimization turned the Spike sequence into a dsRNA mess.
So how is Pfizer measuring their dsRNA? If I had to guess, they will have found the least sensitive assay for the job as they just made a dsRNA nightmare with their codon-optimizations.
Many thanks to Janci Lindsay for forwarding these papers. These point to an antibody driven ELISA assay that targets a 400bp dsRNA. This is a long dsRNA and may not be a good proxy for these modRNAs that are more Rod shaped and have bubbles/stem loops like Viroids.
What is important to recognize is that they are trying to measure dsRNA to understand if the host cells native RNaseIII enzymes (DICER1) will interact with these RNAs.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7314518/
These papers cite a dsRNA ELISA assay from 1991 (Schonborn et al). You really have to dig into these papers to understand the limitations of these dsRNA antibodies. What is important to note is that these assays are documented to NOT work well on Rod-Like viroid dsRNA. This is what the modRNA vaccines most resemble. The PSTVd RNA being measured in Schonborn et al is Potato Spindle Tuber Viroid. You will notice it has many stem loops or bubbles in the dsRNA. You will also notice the dsRNA ELISA assay fails to detect this. This is precisely the type of RNA the vaccine companies are trying to design as its more stable than ssRNA.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC328262/
from Schonborn et al.
While we noticed the GC enrichment and the Quadruplex G enrichment in the vaccines, we hadn’t quite appreciated that the Rod-shapes they were delivering were deliberate.
Zhang et al explains how to do this.
The Zhang paper is attempting to alter the codon sequences to make the modRNAs more viroid like. But this is the exact type of RNA their dsRNA ELISA assay is documented to not detect. Pretty nice trick!
So the codon optimizations are stuck between a rock and a hard place. They don’t want ssRNA as those are more prone to degradation. In order for them to have a stable RNA they need to codon optimize the ssRNA to form dsRNA. This makes the RNA more stable but also a more likely substrate for double stranded specific RNaseIII found in human DICER1 and their stability requirements are in direct conflict with triggering the RNA interference pathway.
So why don’t they just measure what RNaseIII does to their mRNA? This would be a better proxy for what DICER1 is likely to do to this RNA once transfected into a cell. This is the pathway they are attempting to avoid triggering and is heavily underscored in the EMA documents as a concern.
The reason they don’t measure with RNaseIII directly is because if they did this more logical and direct measurement they would reveal a horror show of 42-52% of the vaccine mRNA decaying in the presence of RNaseIII.
This implies orders of magnitude higher dsRNA in the vaccines than is allowed.
Methods
This chart was generated using AccuBlue (Biotum) Broad Range RNA quantitation kit.
20ul of Vaccine is boiled with 2ul of 10% TritonX (~1% final) at 92C for 8minutes. Results are shown with and without the addition of TritonX. 10ul of this boil is placed into 190ul of AccuBlue dye for measurement on a Qubit Fluorometer. 20ul of RNaseIII buffer is added and remeasured to understand the impact of the restriction enzyme buffer on fluorometric measurements. This measurement was repeated as we were surprised to see this much of an impact on fluorescence from the buffer alone but its an important control. 2ul of ShortCut RNaseIII (NEB) is added to the AccuBlue sample and incubated at 37C for 20minutes. Samples are remeasured on a fluorometer. This step is repeated to see if more enzyme can further digest the the RNA. Keep in mind, RNaseIII digests RNA into 21bp double stranded siRNAs. This siRNA will still stain with AccuBlue so the reduction in signal is not expected to go to zero.
Finally, 2ul of Monarch RNaseA (NEB) is incubated at 37C for 30minutes to evaluate if the remainder of the RNA is digestable with this ssRNA nuclease.
Venice.ai is a good source for understanding what these various RNases do and which templates they are active on.
Conclusions
The dsRNA ELISA being used to monitor the dsRNA is in conflict with the stated goals of mRNA codon optimization. This reagent is documented to fail to detect PSTVd RNA which shares the secondary structure of the codon optimized mRNA vaccines: Rod shaped and optimized for dsRNA features while having interspaced stem loops. Reducing the dsRNA will come at the cost of mRNA stability and too much dsRNA and you trigger the TLR3, RIG-1, and the entire RNA interference pathway.
One has to question, why they don’t just inject the protein and skip all this mayhem with RNA transfection? The obvious answer to this question is that ModRNA’s give them the platform which can be rubber stamped as they swap out RNAs for RSV and Influenza. In fact, this rubber stamping just occurred this week as they recklessly approved RSV modRNA without a public hearing on it. They assume the entire platform is modular and you can simply change the sequence and never have to test for safety again.
It appears this RNA platform is some form of quantum paradox. Their papers and patents speak to the importance of codon optimization for dsRNA formation and stability, while their other papers and patents speak to the importance of eliminating the dsRNA.
This is a clear contradiction that requires a forked tongue to verbalize. How much dsRNA transfection can your cells tolerate? Looks like we are finding out in real time.
The ShortCut RNaseIII reaction buffer is below.
50 mM Tris-HCl
1 mM DTT
50 mM NaCl
(pH 7.5 @ 25°C)